The phage field is growing exponentially. More labs are getting funding, and more non-phage labs are moving into the phage field. We decided to run the State of Phage survey as a way to get a glimpse of what everyone is working on, and to see if there were any outliers and surprises in the phages and strains researchers are collecting.
In Part One, we explored the demographics of the researchers who collect phages and host bacteria.
In Part Two, we explored the surprisingly wide diversity of bacteria that phage labs were collecting.
In this third installment of our State of Phage 2020 Survey Snapshot series, we’re taking a look at the different kinds of phages that labs have been collecting. We’ll also briefly compare the phage coverage against the bacterial host strains the labs have been collecting.
Previous issues in this series
State of Phage #1: A wide diversity of backgrounds & phage experience
State of Phage #2: A wide diversity of strains
When we were putting this issue together, there were quite a few surprises that jumped out at us. We hope that you might glean some insights into the phage community at large, and perhaps even get some ideas and inspiration out of our snapshots.
Some caveats before we begin
Our survey is self-reported and importantly only approximates the phage field, so please take all insights we present with a grain of salt. The data is not statistically significant, and is highly biased towards Phage Directory’s reach. With that said, we can still glean lots of insights based on what we’ve collected!
This data set includes 136 different labs. Thank you to everyone who participated in filling in our lengthy survey!
The data has been cleaned to our best efforts, but we understand much more can be done. If you have any questions about the data, or if you are interested in collaborating and analyzing our survey data, reach out to us at [email protected].
Exploring phages through their hosts
When we explore the phages that labs have collected, we look at phages through the eyes of their bacterial hosts. The way we collect and think about phages is through the bacteria our phages infect. We can’t discover a phage without first having the bacterial strain.
In this issue, we explore the diversity of phages collected through the names of bacterial strains, because we’re looking at phage hosts, and our phages’ ability to lyse those phage hosts.
We also take a look at where in the world these phage collections exist, and how large the phage collections are per bacterial host.
Where are labs getting their phages?
Before we dive into the phage collections themselves, let’s look at where labs are getting their phages, and what they’re doing with their collections.
Most of our labs’ phage collections are comprised of phages the labs isolated by themselves. Almost 100 labs (out of 136 total) responded that they isolate most or all of their phages themselves.
Most of the phages that weren’t isolated directly by the lab were received from collaborators of the lab.
Only a few labs order their phages from a culture collection. Curiously, a handful of labs report that most or all of their labs’ phages are from culture collections.
This probably means that most of the phages reported in this survey should be highly diverse, and possibly highly correlated to the labs’ goals and research questions. This might also mean that researchers find it more interesting to find, isolate, and characterize never-before-seen phages, rather than working with existing model or reference phages from existing culture collections.
Is it better to always be finding shiny new phages, or to better understand a few model phages? It’s been almost 100 years since the original seven T phages were specified. Maybe it’s about time that the set of reference phages grew to include more phages (perhaps covering more bacterial strains)?
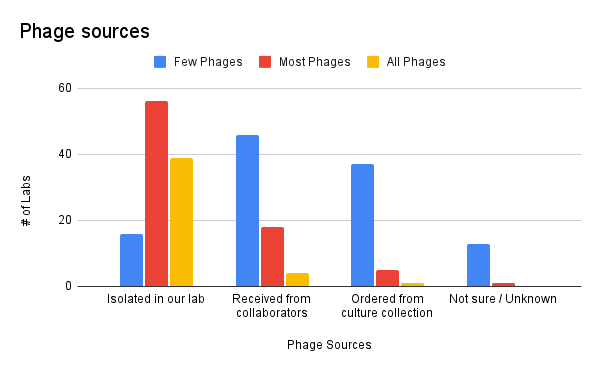
Figure 1. Phage sources (where did you get your phages?).
Where were the phages isolated from?
Most of our respondents’ phages seem to have been isolated from the environment. A small cohort of labs isolated their phages mainly from clinical and animal sources. We’d be curious to dig deeper to understand if phage collections isolated from clinical and animal samples are more “effective” or have “better coverage” against pathogenic isolates than collections mainly comprised of environmental samples.
It’s also curious that the cohort of labs who indicate their phages come from “unknown” sample sources (e.g. environmental, clinical) seem to somewhat map to the group of labs that isn’t sure of where they got those phages from (isolated themselves, got from culture collection, etc). Maybe this means phages were mislabeled as they were stored in the labs, or passed down between collaborators and over time the records were lost.
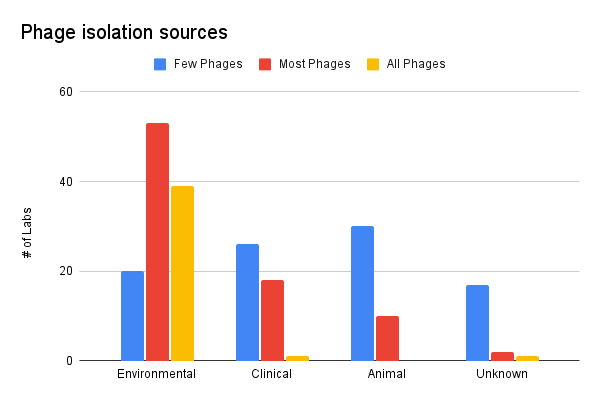
Figure 2. Phage isolation sources (where did your phages come from?).
What are labs doing with their collections?
Most labs (82%) reported that they are actively looking to expand their phage collections by isolating phages on their own. Only a small number of labs are actively getting and depositing their phages into phage repositories like ATCC or NCTC.
Like the above, this implies that the phages that labs are acquiring are likely to be novel and previously uncharacterized. This might also imply that labs are spending significant efforts to isolate, characterize and understand new phages.
| Phage Collection Activities |
# |
% |
| Expanding phage collection |
96 |
82% |
| Isolating new phage |
102 |
87% |
| Acquiring phages from phage repositories |
24 |
21% |
| Depositing phages into phage repositories |
15 |
13% |
| Total responses |
117 |
100% |
The following chart shows a visual breakdown of what labs are doing with their phages. The majority of respondents are only looking to expand their phage collection through isolating new ones. Even though labs are isolating and discovering new phages, very few of those new phages are being deposited into phage repositories.
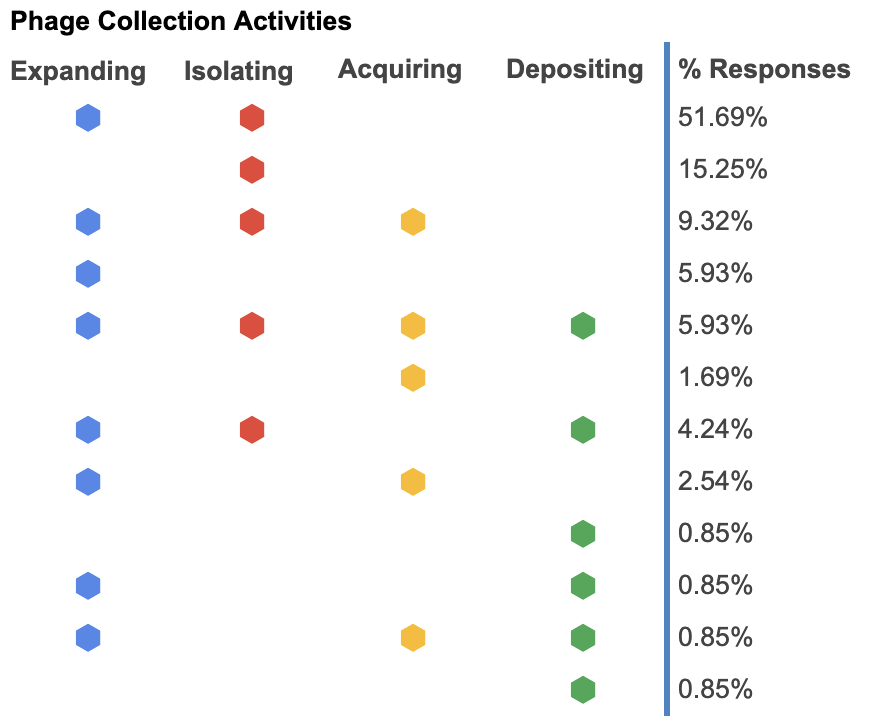
Figure 3. Phage collection activities (what are you currently doing in your lab?).
Which phages are being collected?
We found that labs collected phages against 51 genera, with Escherichia being the most common genus. This group includes reported strains like ExPEC or O157:H7, but for sake of comparison, the strains have been normalized per genus.
It’s not too surprising that there were many phage collections that report having more than 50 phages for Escherichia, Pseudomonas, Klebsiella, or Salmonella. But what is more than surprising is that there are groups with more than 50 phages for difficult hosts like Acinetobacter, *Clostridium, Burkholderia, *and even the notoriously annoying Campylobacter.
It’s probably not surprising that phages targeting pathogens seem to be the most commonly collected. But towards the end of the list we can see labs focusing on plant bacteria like Xylella or human gut microbiome bacteria like Bifidobacterium.
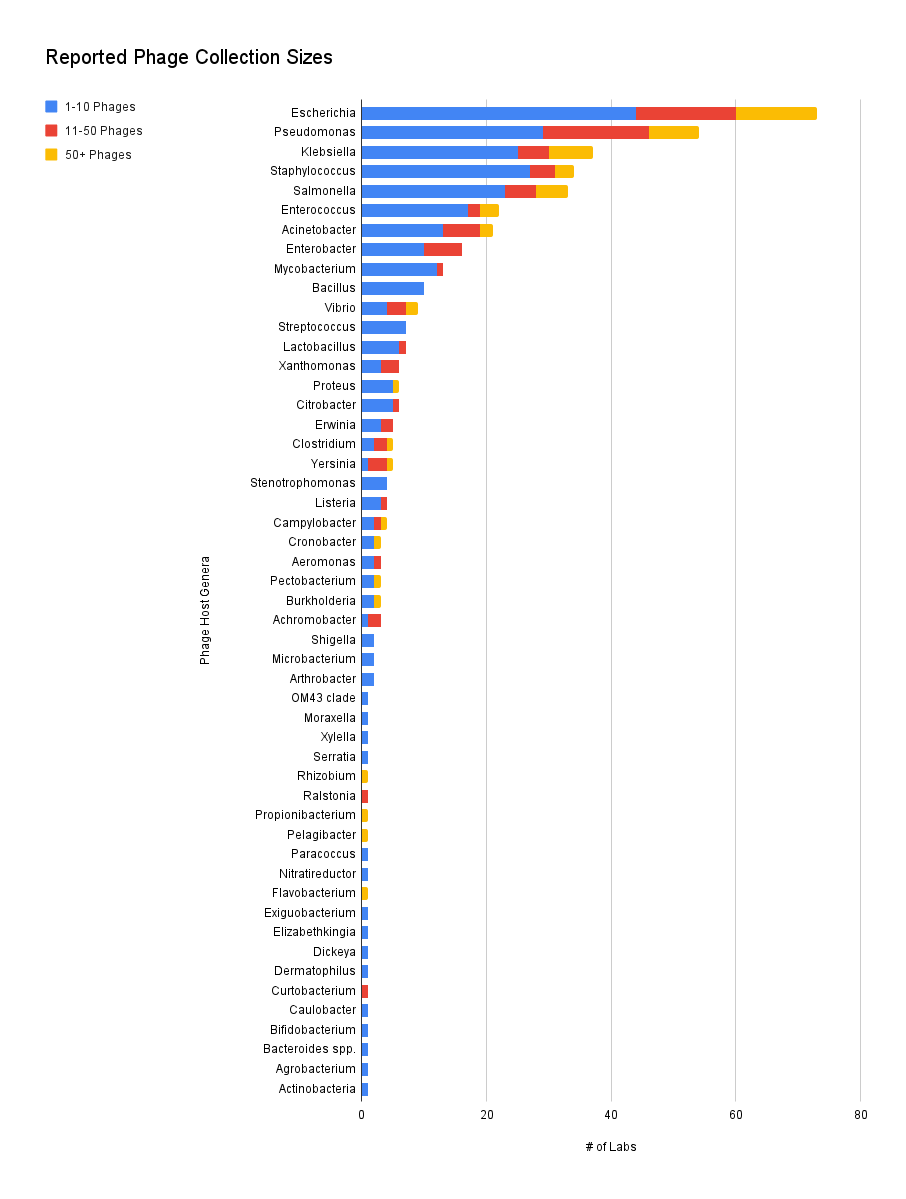
Figure 4. Reported phage collection sizes (Which hosts do your phages target? How many phages do you have against each host?).
How many phages are labs collecting?
When we first devised the survey, we expected most labs to collect fewer than 50 phages. Though indeed roughly 63% of our labs report collecting between 1-50 phages (small to medium sized phage collections), we were surprised to see a large number of large (51-200 phages) to extra-large collections (>200 phages). In the future, we’ll break the question down into more brackets for larger phage collection sizes.
We wonder though — are labs that collect hundreds of phages characterizing and studying each of their phages at the same level of consideration as labs with fewer phages? Are they all equally well-studied and well-characterized? In the future, we should also look at whether large phage collections are based on animal/clinical strains or plant/environmental strains, and dig into the goals of expanding these particularly large phage collections.
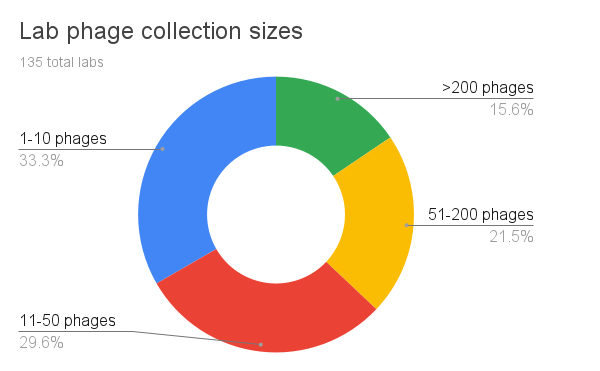
Figure 5. Lab phage collection sizes (how many phages do you have in your collection?).
Where in the world are phages being collected?
In this graph, we count the number of times a phage host is reported, and not the actual number of phages in a collection, per continent. This helps us understand which phage hosts are popular to find phages against in different parts of the world. Just a reminder: our survey responses are heavily skewed towards European and North American respondents, and our number of survey responses does not meet the requirement for statistical significance.
It seems that for the popular hosts like Escherichia, Pseudomonas, Staphylococcus and Salmonella, our responses don’t seem to particularly skew towards any parts of the world. It’s also interesting that Acinetobacter, despite being difficult to find phages against, doesn’t seem to skew towards any continent. This implies that labs work on phages for popular strains in every continent, but the less studied strains might not be as available in labs around the world.
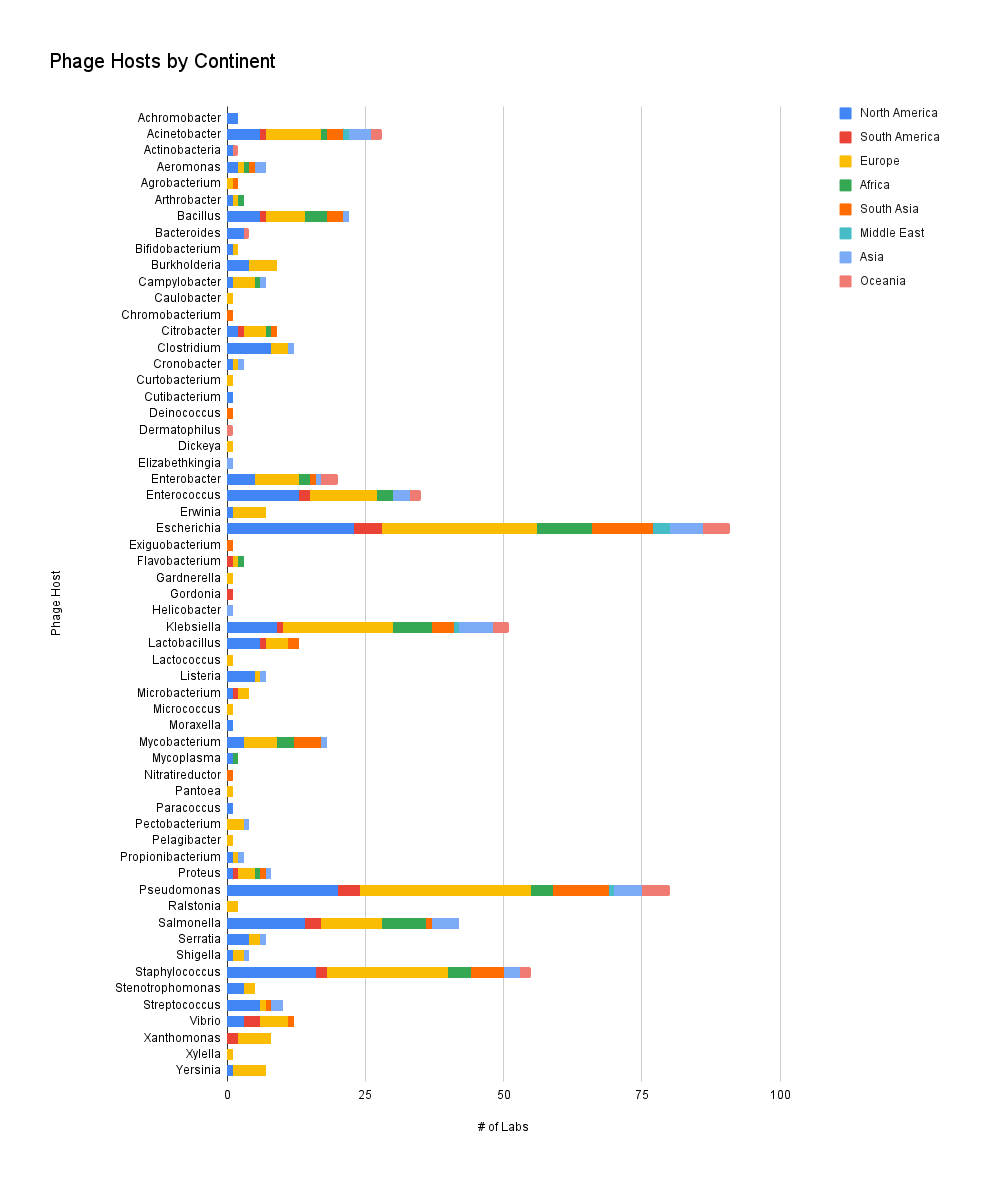
Figure 6. Phage hosts by continent (which hosts do you have phages against? Where is your lab located?).
Where in the world are the biggest phage collections?
Unsurprisingly, our data shows that the biggest phage collections are located in North America and Europe. Partly, this is because our data is skewed towards those two continents, but this also most likely mirrors the amount of funding devoted to phage research each year. We currently lack accurate data to portray the state of phage collections in South America. However, it’s encouraging to see large phage collections in all continents (except Antarctica…).
Our main takeaway from this is that we should be dedicating extra focus on encouraging, supporting and seeking out more phage collecting in places like South America and Middle East.
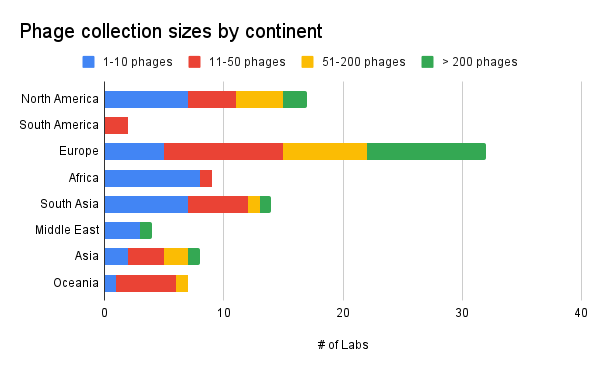
Figure 7. Phage collection sizes by continent (where is your lab located? How many phages do you have in your collection?).
What strains were most difficult to find phages against?
Here’s a word cloud of what people said were difficult hosts to find phages against. We’ve left the responses untouched, so responses like specific strains like VRE or UPEC can stand out.
The difficulty of some of these strains may stem from the inherent difficulty of handling the strains in the lab. One notable exception would appear to be Staphylococcus aureus.
Even though S. aureus is reportedly one of the most difficult hosts to find phages against, the Staphylococcus genus happens to be the 4th most common phage reported collected in our survey (see “Reported Phage Collection Sizes” above). We’ve heard that diverse S. aureus phages are really hard to come by — that most phages are phage K, or very similar to phage K. So this may be why people are reporting it as a difficult phage-baiting host.

Figure 8. Difficult phage hosts (which hosts have you found particularly difficult to find phages against?).
How are labs studying their phages?
The term “characterizing” a phage can mean different things to different labs, as our survey results show. According to the survey data, most phages are genome-sequenced and analyzed for host range. Some labs reported that they’ve subjected their phages to microscopy, burst size, bioinformatics, and stability analysis. Only a few phages appear to have been studied in in vivo models or through antibiotic synergy testing. Very few phages appear to have undergone receptor analysis, proteomics, or transcriptomics. Interestingly, one group reported performing transcriptomics on all their phages. Equally interesting was the group that reports not performing any host range analysis any of their phages. Maybe their collection consists of reference phages (e.g. T4)?
Other free-form entries include: virulence testing, plaque assays, pH stability, temperature stability, ERIC PCR, genetic screens, infection kinetics, genomic DNA extraction and restriction, electron microscopy, and lysis kinetics.
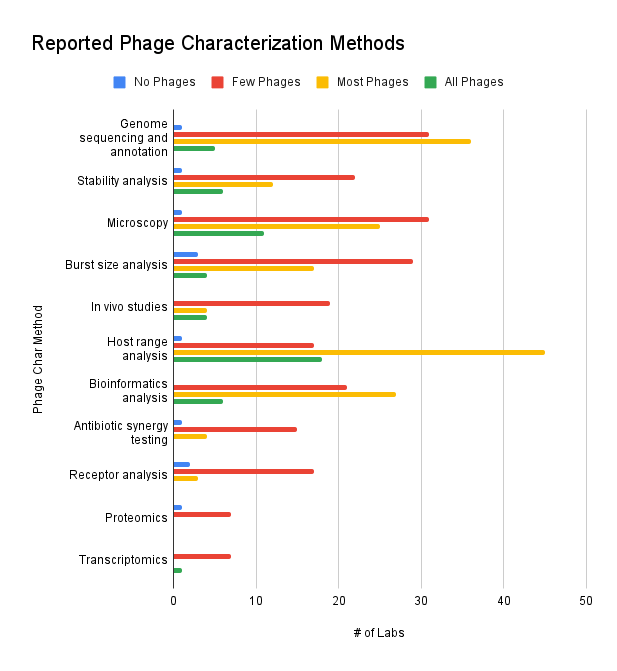
Figure 9. Reported phage characterization methods (which methods do you use to characterize your phages? For each method, how many of your phages have been characterized by this method?).
How common is it for phage collectors to sequence their phage genomes?
Most labs — 104 of 136 (76%) labs — report that they have sequenced at least a few phages in their collections. For the larger phage collections, most phages are getting sequenced. Those operating large phage collections might intend to sequence all the phages they isolate, but are likely limited by sequencing resources and capacity. Smaller phage collections might lack the resources and capability to sequence their phages. The takeaway is that not all phages are sequenced, regardless of the size of the collection.
One surprising outlier is the lab that reports having more than 200 phages but haven’t sequenced any of their phages.

Figure 10. Phages sequenced vs. phage collection size (how many phages do you have in your collection? How many of your phages have been genome-sequenced?).
How extensively do phage collectors characterize their phages?
This chart shows how much each lab considers their phages “fully characterized” according to their own standards. There is a range of levels of characterization that was reported, for all sizes of collections, and comparison between this figure and the one above suggest that there’s still quite a difference between phages that are fully characterized and phages which are merely genome-sequenced.
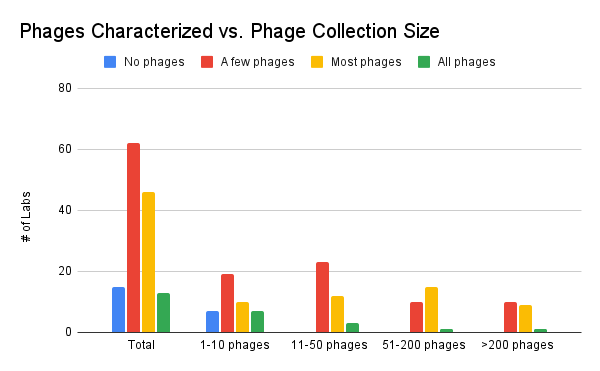
Figure 11. Phages characterized vs. phage collection size (how many phages do you have in your collection? How many of your phages do you consider characterized?)
How do phage collection sizes relate to strain collection sizes and to total target host coverage?
We next wanted to determine how phage (and strain) collection sizes might correlate with the number of strains that labs are both collecting and able to target with their phages. In other words, do larger phage collectors always have large bacterial strain collections, and do larger phage collectors correlate with larger numbers of unique hosts targeted? In the following graphic, we compare the size of the phage collection (grouping labs into small, medium, large and extra-large phage collectors), against the number of unique strains collected and targeted by these groups’ phages.
“Unique hosts” are counted as uniquely reported hosts — this means that in the data, ExPEC is counted separately from “E. coli”.
In the group of collections with the smallest phage collection sizes (1-10 phages), most labs also collect relatively fewer bacterial strains. There are a handful of exceptions, ie. labs that collect more than 50 bacterial strains, but have relatively small phage collections. Interestingly, the group of 43 labs with small phage collections collectively cover 40 unique phage hosts! Though a few labs with small phage collections collect phages for more than 5 different hosts, most seem to be collecting phages against 1-3 hosts.
On the other end of the spectrum, larger phage collections seem to also have larger strain collections. These labs might include culture collections and biobanks. In the 51-200 phage (large) collection category, the labs seem to be collecting a large number of phages across a small number of different phage hosts.
Interestingly, we see from the data that a large number of smaller phage collections combined seems to be able to approximate the total phage host coverage of large phage collections. This indicates that an increase in unique phage host coverage (from a ‘whole field’ perspective) could be achieved through growing and spreading out the number of smaller phage collections, rather than necessarily needing to involve large phage collections at any one center.
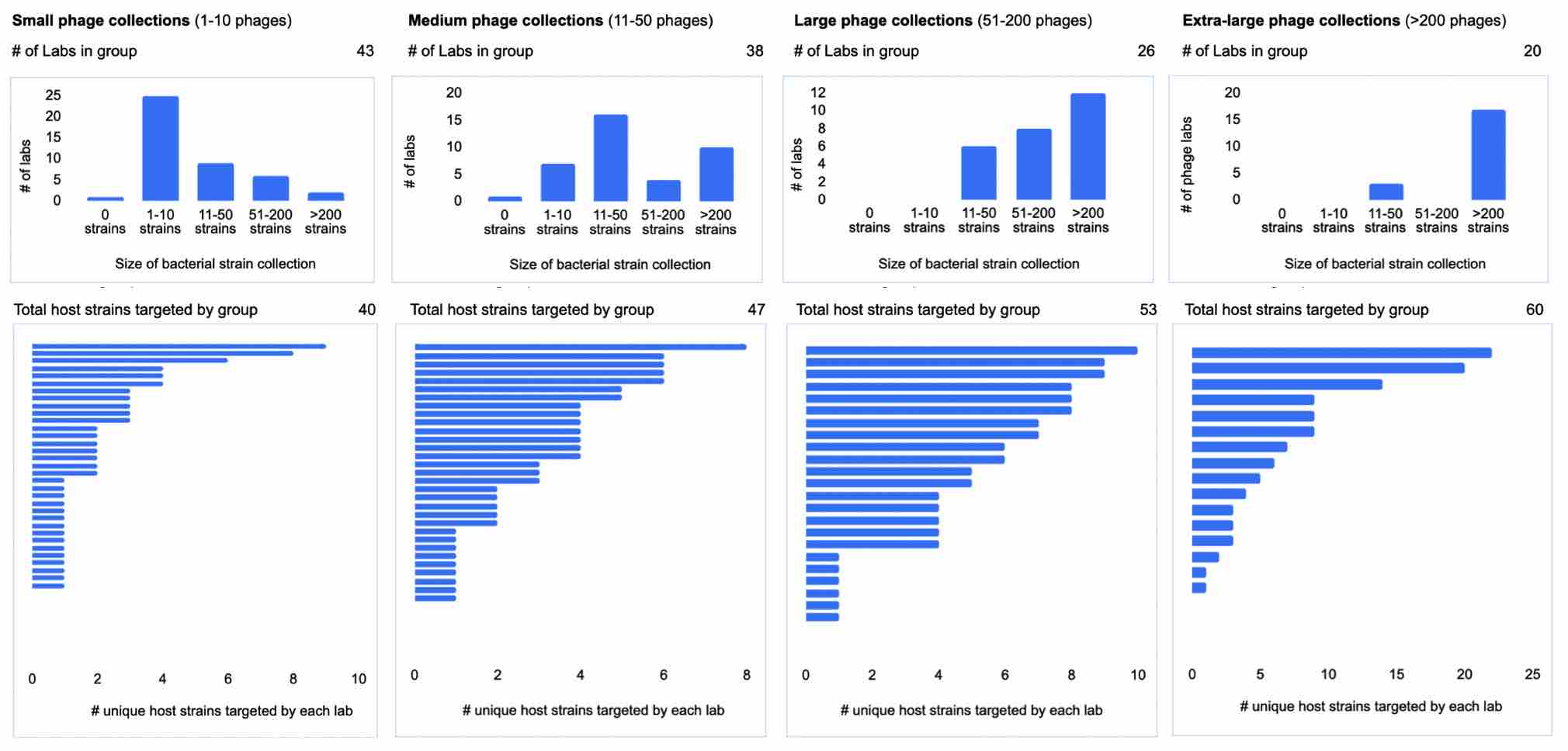
Figure 12. Size of phage collection (small, medium, large, extra-large) vs. number of bacterial strains collected and number of unique hosts targeted (first collectively by group, and then broken down for each individual lab). (How many phages do you have in your lab? How many bacterial strains do you have in your lab? How many strains do you have phages against?).
Of the bacterial strains labs collect, how many do labs have phages against?
Out of the 57 different genera of bacterial strains collected among the labs, 51 (89.5%) of those strains have corresponding phages reported by respondents. The strains that don’t have corresponding phages are marked in blue in the table below. Interestingly, a few phage hosts were reported, but their corresponding strains were not (marked in yellow). One exception in this stems from data cleanup errors, as Cutibacterium and Propionibacterium are both the same genus.
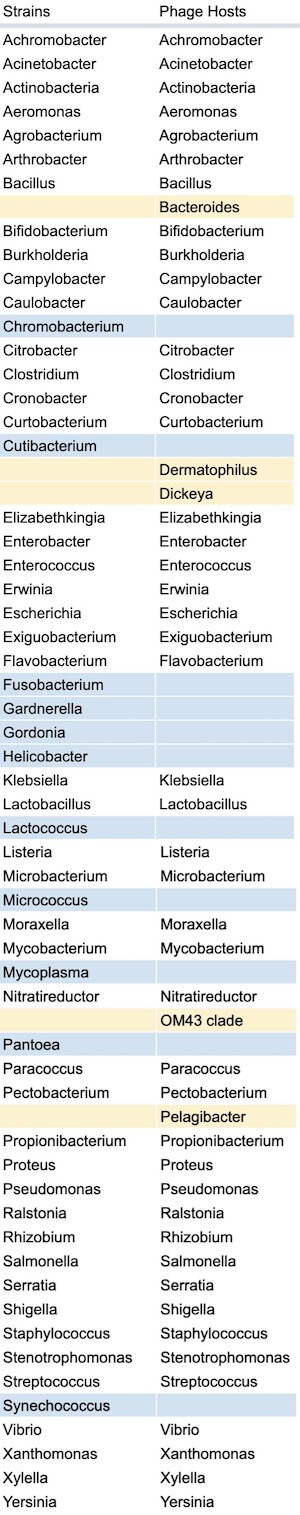
Figure 13. Complete list of strains collected vs. phage hosts collected (which bacterial strains do you have in your lab? Which strains do you have phages against?).
Final Remarks
In this issue, we saw a surprisingly wide coverage of phages across both large and small phage collections. Most labs are isolating their phages themselves, with few getting phages from culture collections (and few depositing their phages).
What struck us as remarkable was that 40 labs with fewer than 10 phages each, had 2/3 of the coverage of 20 labs with more than 200 phages each. Potentially, this means that a wide network of smaller phage collections, each collecting fewer than 50 phages, spread around the world, could conceivably maintain coverage equal to that of large, well-funded phage banks.
Also surprising was the diversity of phage hosts, covering everything from clinically associated, pathogenic hosts to plant, soil, and environmental hosts. We saw phage collections of all sizes on nearly all continents, highlighting that anywhere you look, phages against all kinds of strains can and have been found, and are being studied.
It was also very interesting to see the ways labs are characterizing their phages, with sequencing and host range analysis being (unsurprisingly) popular. Beyond this, it was interesting to see several other methods being used on portions of labs’ phage collections, indicating that phage characterization is being done creatively and extensively around the world, with many labs digging deeper into understanding their phages in their own ways.
We also thought it was interesting to look at what proportions of phage collections have sequenced their phages. As sequencing becomes cheaper, and as more funding continues to flow toward phage research, we’ll soon likely see a massive influx of better characterized, more diverse phages, isolated from more places around the world.
The phage community is nascent but exploding, and it’s really exciting to be a part of this quickly growing field!
How we generated the data
Jan used Observable to crunch the numbers, and used Google Sheets and Vega-Lite to generate the graphs.
Your feedback is welcome!
What are you surprised by from this issue? Did we miss anything? We’d love to hear your thoughts! Email [email protected] anytime!
Looking ahead to part 4
In the next and final State of Phage Snapshot issue, we’ll take a look at the tools and methods researchers use, and their general perspectives and attitudes towards publishing and sharing phages. We’ll also reflect on the future of the phage research community, where we think the field needs to focus, and the future of the State of Phage survey.
Many thanks to Atif Khan and Stephanie Lynch for finding and summarizing this week’s phage news, jobs and community posts!