I’ve never been particularly fast at reading papers. Usually, just skimming a research paper takes me about an hour. Reading a paper — by trying to understand it “completely” — can take me at least 4-5 hours. And it leaves me exhausted at the end.
Most papers I’ve read cite at least 10+ papers. ~5M papers are published per year. If someone were to “read everything that’s ever written” about a topic, they’d never do anything else. And the worst part is, spending all this time reading a paper, then finding it’s irrelevant.
There are tons of great papers out there, but to cut to the chase, we need to focus only on those that are relevant and worth our time. While Pubmed and Scholar are still invaluable, here are a few new search tools that can help you figure out which rabbit holes are worth going down, and which you can skip.
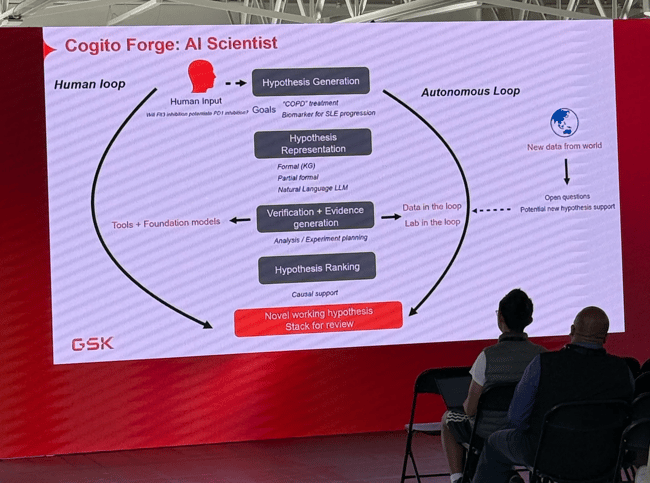
I’m at a GSK talk while writing this, and to my surprise, they’re explaining how they use AI to speed up their paper discovery and hypothesis loop. Ideally, their flow is: Exploration > Literature synthesis > Hypothesis > Update knowledge > More Exploration. With these new tools, they can scale up this process to hundreds of thousands of iterations. They’ll then review the final stack for the most interesting ideas.
While the following tools aren’t at GSK’s level, they can still help your lab approximate this flow. Below are some of the tools that Jess and I like to use for research and reading tasks. (We’re not affiliated with or get paid by any of them — we’re just fans!)
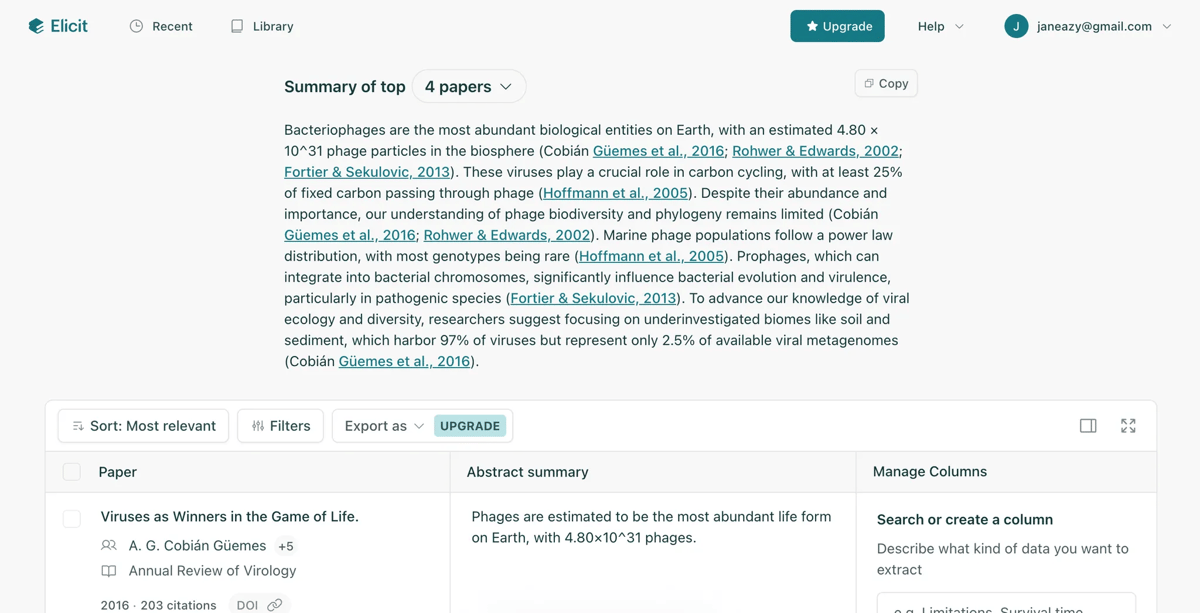
Elicit.com is designed for researchers, and generates a really nice table of all papers relevant to the search term. Jess and I have been Elicit fans for a very long time!
Science-specific search tools
We’ve all used Google, Google Scholar, Pubmed, and Mendeley for regular research work, but here is a selection of new search systems specifically geared for scientific research. Think of these as chimeras of Google Search, Mendeley and other science platforms. These platforms are meant for performing an array of tasks like lit reviews, data extraction, and full research tasks.
Elicit - http://elicit.com - I’ve spent quite a bit of time with Elicit, and this is my favorite tool for digging deep into one specific topic (e.g. phages and optogenetics). It searches a graph representation of related papers, and can summarize an entire thread or domain fairly quickly, surfacing exactly the papers you need to know.
SciSpace - free + paid tiers - https://scispace.com - Similar to Elicit, SciSpace is a system for searching for papers along queries and topics. It also doubles as a powerful reference manager, and has AI features like summarization. I commonly will check results between Elicit and SciSpace.
Consensus.app - free + paid tiers - https://consensus.app - Consensus is a clever take on searching science. Designed more for laypeople, it takes statements like “does phage therapy work” shows search results both for and against the question, and figures out where the consensus lies. This one is excellent for writing more accessible articles for science communication and similar tasks.
FutureHouse Platform - free - http://platform.futurehouse.org - FutureHouse is a nonprofit for building science automation tools. Their new Platform is a framework for comprehensive lit search, critical analyses, and hypothesis validation. Eventually, they’re aiming to build a full “automated scientist” where they can pair up both data and lab in a closed loop. Learn more about their work here.
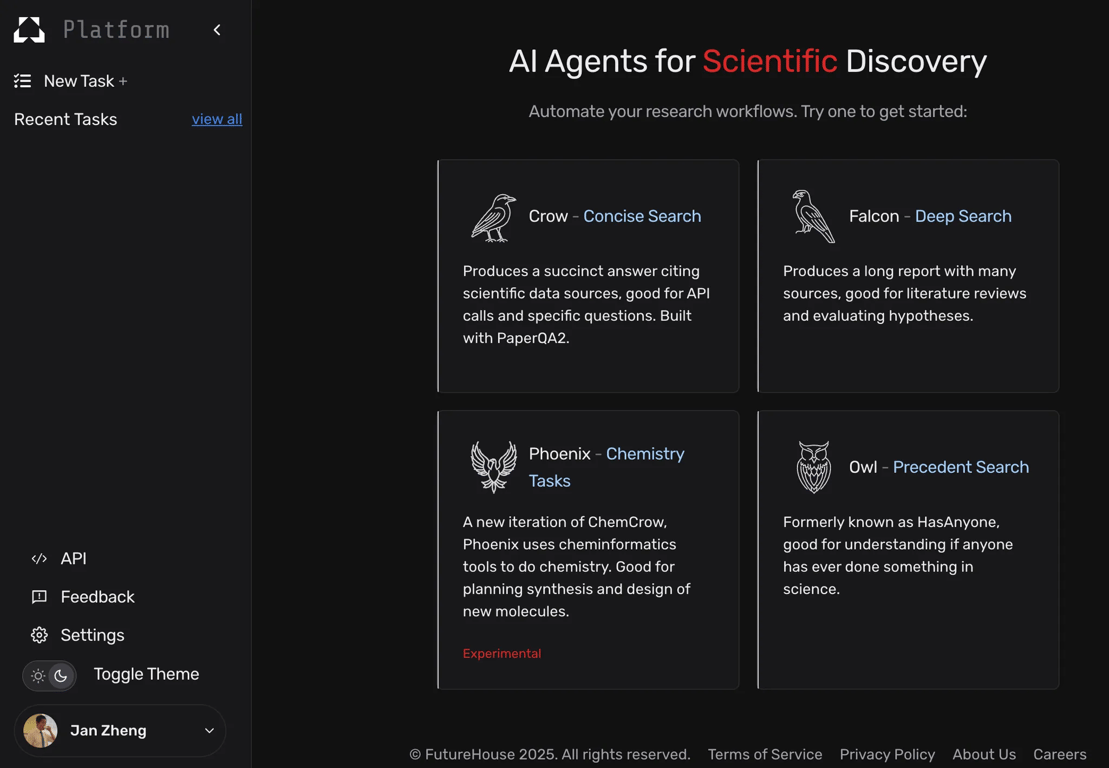
The Future House Platform has various nuanced search systems for researchers. https://platform.futurehouse.org
General search tools
There’s also quite a few AI-native search platforms that have been released in the last few months. These systems summarize, contextualize, and extract data from hundreds of search results. Below is a short list of the search tools that I’ve tried.
Kagi Search - free + paid tiers - https://kagi.com - Kagi is not an AI search tool. It’s an old-school query-based search like Google, but unlike Google it doesn’t have ads. Since they’re not incentivized to show sponsored results, the results are generally much better than what Google shows. They do have their own version of Deep Research though, but it’s still in beta.
Google Gemini Deep Research - free + paid tiers - gemini.google.com - Invented the concept of Deep Research. Runs dozens to hundreds of Google searches, finds relevant papers and links, and cites the sources as it answers the user’s questions. This is very good for terms that are otherwise hard to find on Google, but can take several minutes. You can also look up papers related to a given paper, or do a quick check if anyone has ever thought of a specific hypothesis. This alone will save you many hours a week.
OpenAI Deep Research - free + paid tiers - openai.com - Very similar to Google’s Deep Research, and works somewhat similarly. I think they use Bing Search instead, but otherwise the results are similar.
Perplexity - free + paid tiers - https://perplexity.ai - This is a completely new search service, and can sometimes have better integrations with Pubmed and other paper databases. They have standard search and deep research, as well as a feature that creates “wikipedia”-like pages of results. I find their search is faster, and seeing how it searches for various papers, “thinks” about the results, then continues to search, is really neat. I normally use Perplexity as an alternative to Google.
Anthropic Claude Search - paid - https://claude.ai Claude is an underdog in the AI field, and excels at writing prose and code. We use Claude often for paper summarization. Claude Search is a new feature, and while it doesn’t give as detailed reports as Gemini or OpenAI, it does create better writing outputs.
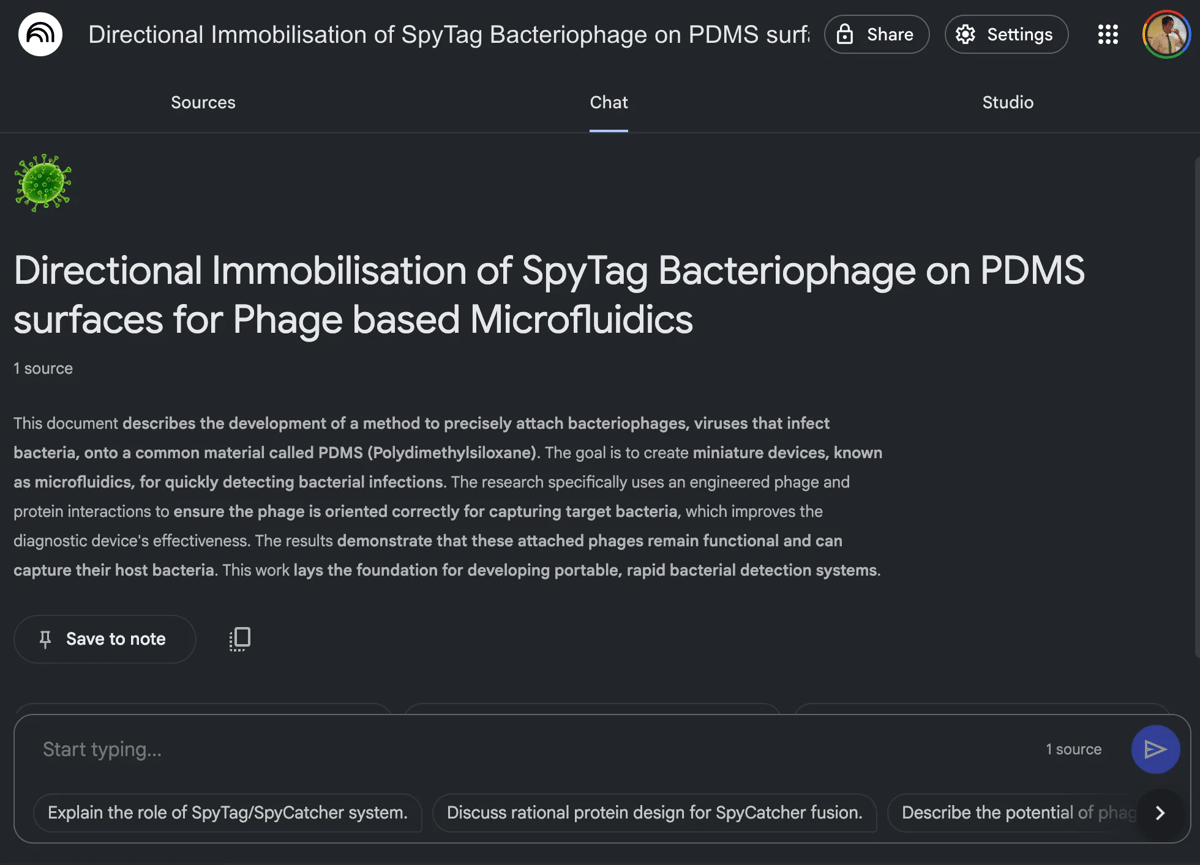
NotebookLM offers a way to chat with papers, plus you can use it to create engaging podcasts! notebooklm.google.com
Audio research tools
Listening to a paper quickly helps me get the gist of a paper, and helps me find all the parts I should pay attention to, before sitting down and diving deep into it. I highly recommend listening to any paper you’re seriously considering fully understanding.
NotebookLM - free + paid tiers - https://notebooklm.google.com - Creates a podcast from (and lets you chat with) any paper, PDF, or report. The ~10 minute podcast is great for quickly grasping papers from areas I’m not too familiar with (e.g. machine learning)
Listening - $13/mo - https://listening.com - Creates a full narrated version of any paper. Jessica uses (and pays for) this one. The audio isn’t perfect, it doesn’t handle images or tables, and sometimes it breaks in funny ways. But it’s still great for long commutes.
Groq’s experimental Deep Research, Fast, which creates a research report in ~10-15 seconds https://deep-research-fast.vercel.app
Programmable Search Tools
For researchers and bioinformaticians who write code, there are a few other notable tools that can mostly only be accessed programmatically.
Exa - Free + paid tiers - https://exa.ai - Can run multiple, parallel searches, and can be useful for those building a research summarization tool. The Websets feature does research in the background and can dig up thousands of relevant papers and other sources on a topic.
Jina Reader - Free + paid tiers - https://jina.ai/reader - Is very good at reading and taking screenshots of websites, but can struggle with research sites. While it can’t access paid journals like Nature, it can access most open source journals.
Groq Compound - Free + paid tiers - https://console.groq.com/docs/agentic-tooling/compound-beta - lets you build advanced web search and code execution through a single API call. This makes it easy to build your own custom versions of Deep Research and AI Scientists, customized to your own needs and your own data. (Side note: this is where I work now!)
Bonus tool: a super fast version of Deep Research, powered by Groq: https://deep-research-fast.vercel.app
One last note
That’s a lot of tools for search!
Whichever option is better, is completely subjective! Since they all have a free tier, there’s no excuse not to try a new one every week, and then compare for yourself. Given how large the potential benefit is of trying one of these tools, it’s really worth the 30 minutes to try them.
These tools are free, they can shave off countless hours of research, and they are free to start, so there isn’t really any excuse to not try them.
Happy reading!
~ Jan











