“Among the many lessons of COVID-19, one of the most profound is this: You don’t need to be big to be powerful. You just need to be infectious”
Chris Anderson, Curator of TED Conferences writes this in the introduction of his recently published book, Infectious Generosity. He goes on to say, “I want to persuade you that one such possible contagion could actually transform the world for the better. Its name?”
Phage!
Actually, he writes “generosity”. His message is that many of the same things that we see amplifying social pathogenicity and entropy (social media and generative AI, for example), can be leveraged to restore a healthier balance to our systems when we tap into our real shared values. Generosity can “go viral” as a powerful countermeasure against fear and hopelessness.
In getting to know the phage community, I see that this spirit of infectious generosity and cooperation is strong, healthy, and needed. The beneficial infectiousness of bacteriophage can similarly empower us to keep in check disease spread and untreatable infections from pathogenic viruses and microbes.
Capsid & Tail readers are keenly aware of how phage therapy can combat bacterial pathogens when billion dollar drugs fail. The phage community has come together to use incredibly powerful biology pro bono, because the system of pharmaceutical interventions we have relied on is failing. A new system is needed for One Health that brings together an integrated set of tools, including education, communication, surveillance, prevention, and treatment. Vaccines are another important component of that system, and phage can empower us all to produce them.
The Simple Epidemic-Ending Defense (SEED) System
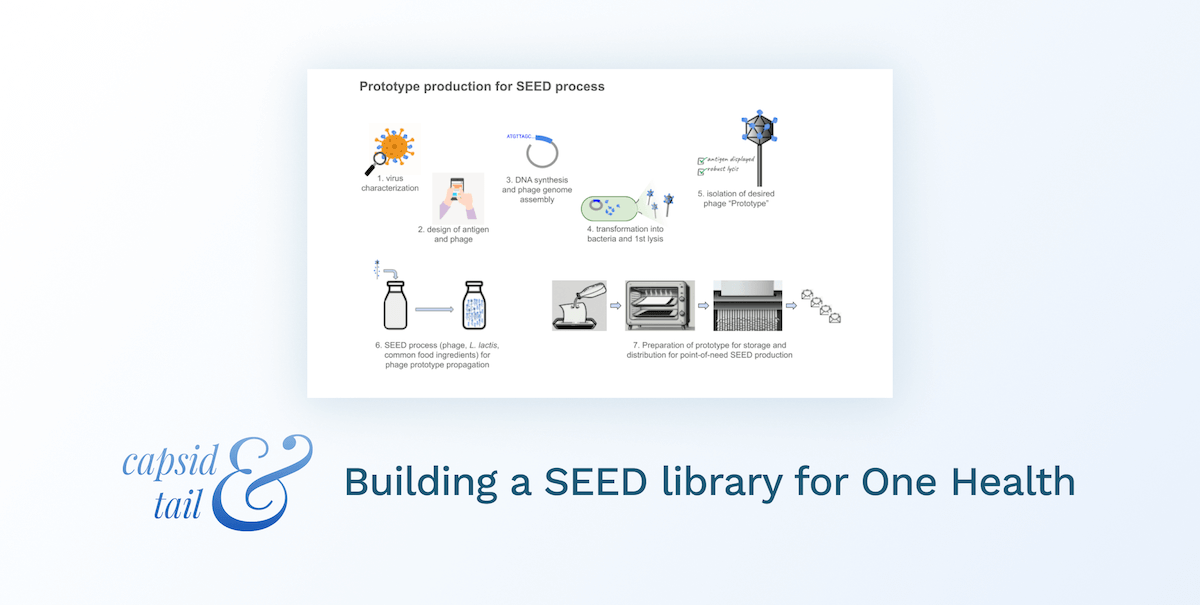
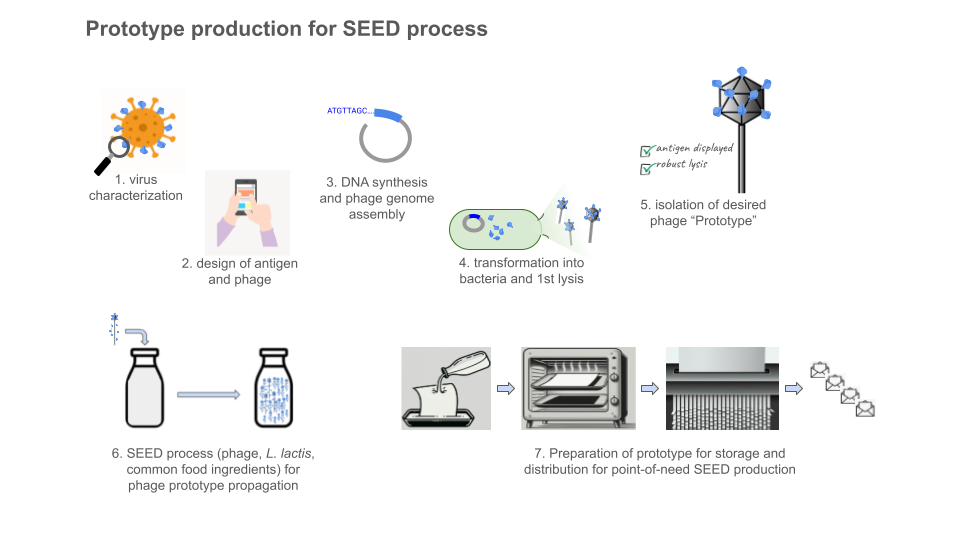
The SEED system was conceived of and designed to democratize vaccine production. Greater San Diego Biological Solutions (GSD Bio) was founded to build the SEED system as a public good. It is a lactococcal phage-vectored vaccine production platform and its purpose is to prevent epidemics.
Vaccination protects our health and lives and vaccine development technology has been improving rapidly. Access to vaccinations, critical to preventing epidemics, has not kept pace. The COVID-19 pandemic thrust this into my awareness. Despite living in the US and having good access to healthcare, our system showed its failure points and we felt the fear of “no one is coming to save you” (another take-home message from this pandemic, articulated by Charity Dean in The Premonition). I realized this was a massive global access problem, and far more serious than what I was seeing around me. I was activated to dedicate what experience, knowledge, and resources I had to working on this problem.
The initial vaccines we set out to produce were against SARS-CoV-2. Upon learning more about the growing problem of the highly pathogenic avian influenza (H5N1) panzootic, we expanded our initial designs to include those that could be used to fight the most current H5N1 viral spread.
The SEED platform employs Lactococcus lactis (unmodified, used in the making of cheese, sauerkraut, kimchi, etc), which can be cultured in simple food-ingredient media. A lytic phage (specific for the subspecies of L. lactis used) is modified to encode a protein of interest (such as an antigen for vaccine). The bacterial cells are infected by these phages and their biosynthetic machinery is repurposed to produce many more phage particles, along with the protein antigen.
Where phages for phage therapy are generally propagated with the pathogenic bacteria they are meant to target, SEED phage are specific to probiotic bacteria and the resulting lysate does not contain pathogens or endotoxins (the entire process is designed to avoid any pathogens or toxins at any stage). Because of this, purification and downstream processing steps, which usually require special supplies, equipment, and skills, are not necessary. The lysate can be simply administered as a mucosal vaccine and is versatile to fit the need.
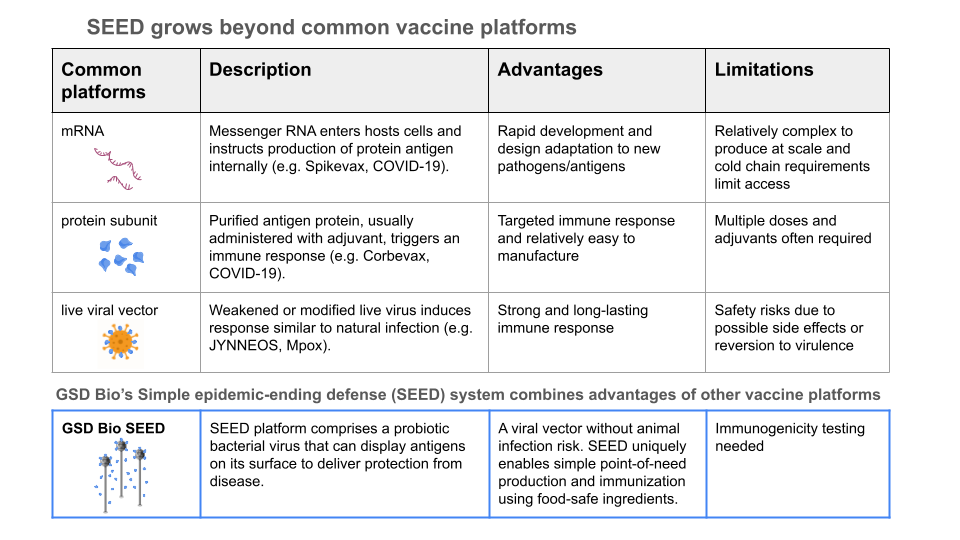
The initial SEED design produced antigen protein in the lysate as a protein subunit vaccine. Newer designs incorporate translational fusion of antigens to phage structural components (see the table above comparing the SEED platform with conventional examples, and dive deeper at gsdbio.com). This can be thought of like phage display of antigens (when display is desirable), but I think of it more as a live viral vector vaccine, where the virus is a phage that can only replicate (wherever needed) and self-assemble into the desired lysate vaccine when it has L. lactis to infect. Traditionally phage display is done in the lab or manufacturing facility with (typically coli-) phage which must be processed, purified, formulated and administered as a vaccine dose (when used as a vaccine or therapeutic, instead of as a screening tool).
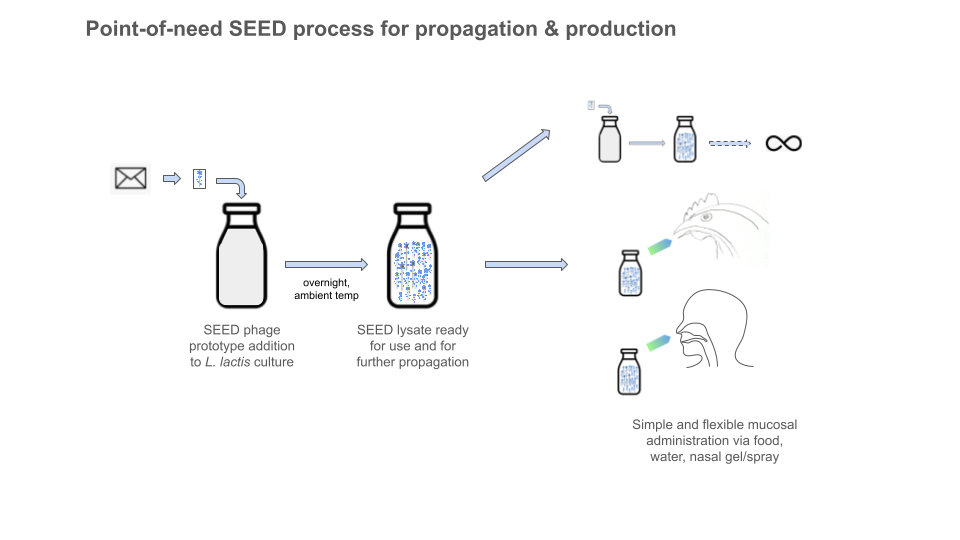
In this way, the system is designed to mimic a natural infection, but with friendly surrogates and without any pathogens, toxins, or downstream GMP biomanufacturing / purification / formulation infrastructure required. The end user can produce vaccines on-site or in situ, without special tools, skills, or equipment. We are democratizing biologics production by letting biology powerfully handle the most complex (and otherwise supply-chain-dependent / resource-intensive!) parts. For all of us needing access to disease prevention tools in communities across the globe, the process is like making buttermilk.
There are limitations of course, some known and some we’ll learn.
The SEED process does not currently result in high concentrations of peptide or protein in the lysates. For vaccines (and some therapeutic peptides) that may not be a limitation (it can be a good thing) as long as they elicit a strong useful response. Particularly because they can be produced right where and when they are needed and can avoid some of the protein stability challenges faced by other platforms.
Some proteins will be more challenging than others. Some viral antigens, for example, could be more challenging to make properly with a bacterial system if there are specific post-translational modification/maturation/folding requirements. While important in antigenic changes involved in evasion of adaptive immunity, I wonder how important these actually are for effective vaccination. It has been shown that viral antigen display (on other viruses and virus-like particles, for instance) and peptidoglycan matrices (GEMs) can stimulate the innate immune response pathways needed to ultimately promote adaptive immunity.
The protein antigens themselves are processed into short peptides for the T cell presentation required for humoral and cellular immunity. There are additional encoded adjuvants that could be co-expressed in the SEED system if needed, but we would prefer to keep things as simple as possible. Furthermore, there are many diseases to protect against, and many antigens that could be produced and tested against each. Particularly if they could be made rapidly and at low-cost. We would start where the platform could be most useful the soonest. There are many smart people designing and screening (potentially) effective vaccine antigens. We want to
remove the gap between that upstream design and administered mucosal vaccinations for global health access.
The maximum size of the payload (the sequence of DNA we can insert into the ~30 kbp phage genome) is yet to be determined. So far, we have done ~3.5 kbp, but we suspect one could go to around 5-10 kbp. This may not be worth focusing resources on. Pushing the upper limit will likely result in less consistent success. Again, we prioritize simplicity and reliability. If we want to multiplex (as for multiple antigen vaccines against bacterial diseases), there are strategies to combine multiple phage prototypes.
The phage should be able to replicate in the mucosa (when desirable) if there is L. lactis present to infect. We intentionally designed the system with L. lactis because it is non-colonizing in animals. So L. lactis can grow and exist in the body (it is low pH- and salt-tolerant too) for a while, but it’s a fairly transient situation (days, weeks, when no phage present). So it is not part of the microbiome. Eating it, drinking it, or its nasal application should not disrupt the microbiome, nor will its lysis. The lactococcal phage we use is lytic, meaning it doesn’t integrate into the L. lactis genome (or any others). In fact, the phage’s entire replication cycle (infection, hijacking of bacterial machinery, replication/assembly and lysis) is reportedly as short as 30 min, with a burst size of 130 (new phage particles released per infected cell). When there are no more L. lactis cells to infect, the SEED phage are unable to replicate and should pass out of the animal host. So the whole system is self-assembling and self-clearing. Designed to only leave behind the desired protective immune response.
Which comes to where we are. We know the platform works with regard to making desired proteins as designed. We have validated the construction of the phage prototypes, the extreme stability and simplicity of components, protein production, self assembly/antigen display. We have shown antigenicity of the produced vaccines (designed against coronaviruses and highly pathogenic avian influenza viruses). Antigenicity meaning that the produced antigens bind the relevant diagnostic antibodies in vitro (measured by immunoassays).
Immunogenicity testing of produced vaccines is the next step. We need to learn the degree to which they cause the immune system to mount an effective protective response. Exploring neutralizing antibody production, cell-mediated responses, and ultimately challenge studies, will guide best paths forward. We may incorporate encoded adjuvants (like subunits of bacterial outer membrane proteins) for greater immunogenicity. This is part of the next generation designs we are working on (in case it is needed - but we don’t know yet). Immunogenicity and challenge studies are beyond our capabilities at GSD Bio and we are working hard to join forces with those who can do the appropriate testing.
Joe Warner and I are the operational team at GSD Bio. As Director of R&D, he is in charge of lab work and I have been primarily driving efforts outside of the lab. GSD Bio was founded in November 2022, we started the lab in January 2023, and we have made pretty incredible progress (Joe is terrific). Our Board is Katherine Arnold, Carl Abulencia, and Jazell Estadilla Handke (and me). With different perspectives and experiences, they all share a science background and a strong sense of mission. Additionally, we have been fortunate to sponsor a team of students from the California State University, San Marcos College of Business Administration on their Senior Experience Program project. They are consulting to help GSD Bio build better structures toward awareness and support of the work. GSD Bio is small, resourceful, and unswervingly committed to biology pro bono.

A path to a healthy future: Our next steps
GSD Bio has been self-funded until now. We have done it all with $200,000, but now need to connect with impact-driven supporters to continue our work. It is challenging. There seems to be a lot of risk aversion in philanthropy and less familiarity with small, community-driven research organizations. Most seek something ready for them to carry over the line, provided to them by an organization with proven financial sustainability. Investment (for profit or for good) is biased toward supporting reactive interventions (prevention is incredibly more effective, but not as marketable or visible). The systems (again, for profit and nonprofit) are deeply dependent on competition for limited resources. We have applied to two CEPI (Coalition for Epidemic Preparedness Innovations) grants, a Robert Wood Johnson Foundation grant, and a USDA-APHIS grant. The most recent proposal to CEPI was for an exploratory platform innovation grant, where we were co-applicants with an incredible and veteran team of pro bono vaccine developers to do preclinical studies of the SEED platform-produced coronavirus vaccines. The proposals were not accepted. Our co-applicants remain interested in the partnered studies, but they do not have funding for them either. There is a similar theme with work on feline coronaviruses.
On the science side, the SEED platform is powerful and versatile. We will continue to work to get it out there for public benefit. I know there are people for whom our mission deeply resonates, who will take action with us to collaborate and to support the work. I need to find them.
Phage are small, infectious, and powerful allies in biology pro bono. They provide tremendous tools for research, for fighting otherwise untreatable infections, and now for prevention of disease transmission. The phage community is full of infectious generosity. I sincerely hope the focus remains on cooperative research and on leveraging helpful viruses for health. I hope you will reach out to me at [email protected] with your curiosity and ideas for collaboration.