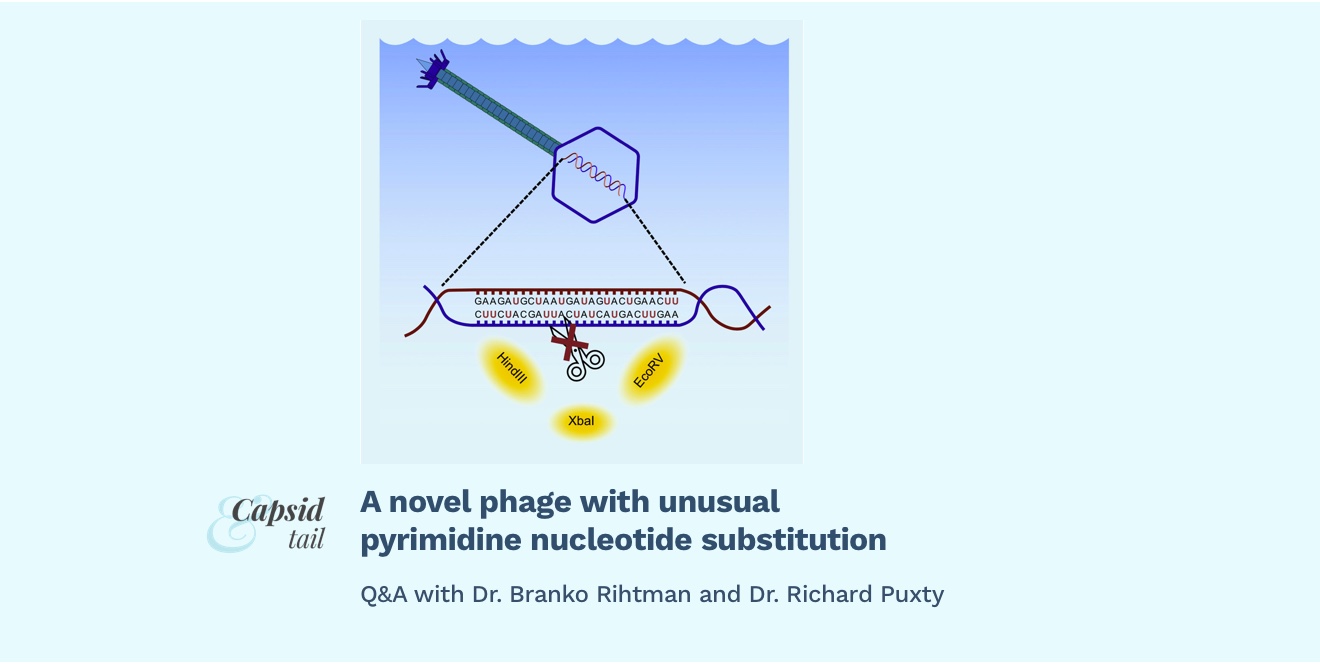
Branko Rihtman and Richard Puxty (University of Warwick) recently published a new paper in Current Biology documenting an unusual nucleotide substitution from deoxythymidine to deoxyuridine in a novel family of phages belonging to Naomiviridae. These substitutions render the phage DNA resistant to digestion by common restriction enzymes such as EcoRV, XbaI and HindIII.
Madhav: Could you give us a little background about yourself and your research interests, to start?
Branko Rihtman: I studied for my undergraduate and MSc degrees at Hebrew University of Jerusalem, Israel, focusing on transcriptional regulation of nitrogen stress in marine cyanobacteria in the lab of Prof. Anton Post. I completed my PhD under supervision of Prof. David Scanlan at the University of Warwick and Prof. Martha Clokie at the University of Leicester, studying the transcriptional landscape of cyanophage infection under phosphate stress conditions, as well as characterising some of the phage auxiliary metabolic genes. Since then, I have worked as a postdoctoral researcher in the lab of Prof. Yin Chen at the University of Warwick, isolating and studying roseophages and their effect on lipid content of roseobacter cells, work which has resulted, among others, in this paper. My main research interest is the presence and function of auxiliary metabolic genes in phage genomes and different ways in which phages affect the metabolism of their host over the course of infection.
Richard Puxty: I am a molecular microbiologist. My early interest was in photosynthesis in prokaryotes, but since a PhD at Warwick (2010-2014), I have become interested in phage biology, predominantly through the discovery that phage genomes encode core genes of photosynthesis. Since then, I have fallen in love with phage research and in particular, the molecular genetic arms race between phage and their hosts.
M: Can you give us a quick summary of your new article?
B & R: In this article, we describe a novel family of roseophages — Naomiviridae — which were isolated from sea water from two very distant locations: water from the canal under the Rialto bridge in Venice and coastal water from Puerto Morelos beach in Mexico. The fact that two very similar phages, which are at the same time so distinct from any other currently known phages, were isolated from such distant locations hints towards global distribution of this viral family.
This observation then raises the question: why were they not previously seen in numerous metagenomic studies? The reason probably lies in the same problem that we encountered when trying to sequence these phages: the library preparation step, which is based on a transposon insertion step, has failed over several attempts.
We have finally succeeded to sequence the genomes by performing an initial whole genome amplification step, but this has alerted us to the fact that these genomes may have some unique modifications that are preventing transposon insertion in the initial steps of the sequencing process.
To our surprise, we found that these phages have completely replaced deoxythymidine in their genomes with deoxyuridine. Genome sequencing of the phages has shown that they possess in their genomes genes encoding some of the enzymes which would potentially be required for this conversion of dTTP to dUTP, such as cytosine deaminase and dTTPase, located in proximity to the viral DNA polymerase. Querying of environmental metagenomes has discovered other viral contigs which contain putative cytosine deaminase and dTTPase orthologues in a similar genetic context to the one found in DSS3_VP1 and DSS3_PM1, pointing to the presence of the same or similar genomic modification in other phages in marine environments.
Finally, uniqueness of these phages’ features, as well as large phylogenetic distance from all other currently known phages, has helped us classify them as members of a novel viral family Naomiviridae, forming a new genus Noahvirus within that family. Hopefully, these results will help inform future metagenomic studies of marine environments and will facilitate identification of additional phages belonging to this viral family.
M: Can you share more about roseophages and their importance?
B & R: Compared to other viral groups, very few roseophages have been isolated and characterised in detail. Since the members of the Roseobacter clade are important participants in the biogeochemistry of the ocean, participating, among other important biogeochemical processes, in the production of methylated amines, which are important precursors in nucleating particles involved in cloud formation above marine environments, thus having a global effect on the climate.
Due to the importance of their bacterial hosts, roseophages also play an important role in these biogeochemical processes by shaping the Roseobacter community dynamics and affecting the relative presence of these bacteria in the ocean. They also influence the flux of dissolved organic and inorganic compounds via the viral shunt of the microbial loop, where lysis of the bacterial host cells releases nutrients into the water and makes them available to other trophic levels of the ecosystem.
In addition to their role in the bacterial life cycle, these phages possess orthologs to bacterial metabolic genes which can provide insight into specific niche adaptation mechanisms and teach us about evolution of bacterial enzymes under nutrient stress conditions, processes which are much more pronounced in phages than in their bacterial hosts due to the preponderance of evolutionary pressures exerted on viruses compared to the bacteria they infect.
Finally, constant arms race between the bacteria and the viruses that infect them have produced a large number of viral defense systems in bacteria, which have been finding their way into different biotechnological applications over the years. Therefore, it is very important to study the novel phage genera, especially those that show novel features and unique adaptations to compete against their host defense mechanisms.
M: How was the nucleotide substitution discovered, and what previous research has been done on such novel nucleotides?
B&R: The nucleotide substitution was discovered due to the fact that the library preparation step of Illumina sequencing has failed a number of times. The only way we could sequence the genomes was if we performed a whole genome amplification step prior to the library prep.
After sequencing the genomes, we were intrigued about the nature of the modifications, and so we sent the purified viral DNA to Dr. Peter Weigele at New England Biolabs in Cambridge, MA, USA. He and his team performed a mass spectrometry analysis of separate nucleotides forming the DNA of Naomivirus phages, and have shown that dTTP is completely absent from the genome, while being replaced by dUTP.
dTTP-to-dUTP genome modification has been shown to be present in Bacillus (Takahasi et al. 1963) and Yersinia phages (Kiljunen et al., 2005), but this is the first time it has been described in marine phages.
M: Could you expand on the inhibition of restriction digestion by common endonucleases, on the UDG endonuclease VIII assay, and on why this assay was chosen?
B&R: We hypothesised that the purpose of the phage genome modification was to protect the phage DNA from host endonucleases, and therefore tried to digest it with some common dUTP-sensitive endonucleases.
Together with endonuclease assay, we performed the UDG-endonuclease VIII assay, in which dUTP is removed from the genome by the action of UDG, creating an abasic site in the genome which is then open for digestion by endonuclease VIII.
We performed these experiments as an additional, independent way to experimentally demonstrate the presence of dUTP in the phage genome, using the whole-genome amplified DNA as a control that shows that sensitivity of native phageDNA to UDG-endonuclease VIII assay is not sequence-dependent.
M: Could you share a few insights into this phage’s genome sequence and lifecycle?
B&R: DSS3_VP1 and DSS3_PM1 belong to the siphoviral morphotype and have a latent period of 110 and 210 minutes respectively. Their capsids possess an icosahedral structure measuring 47–55 nm in diameter, with the tail measuring approximately 230–240 nm in length. One-step infection experiments in liquid culture show that DSS3_VP1 and DSS3_PM1 have infection kinetics that differ from previously reported double-stranded DNA (dsDNA) roseophages, which were shown to not fully lyse the infected bacterial culture, while Naomivirus phages achieved complete lysis of the liquid culture.
From the genome sequence perspective, Naomivirus phages are relatively conventional, apart from the genes required for conversion of dTTP to dUTP. Similarly to quite a few other dsDNA phages, they possess a putative type I self-splicing intron, containing a type VII homing endonuclease within the sequence of the large terminase gene TerL. They also possess both a DNA and an RNA polymerase, which is expected due to the unconventional nature of their genome. Similarly to almost all known sequenced phages, about 80% of the predicted ORFs encode hypothetical proteins with no known orthologues in other genomes.
M: What is the importance of studying these nucleotide substitutions, and how can this impact our understanding of life in nature?
B & R: There are several important aspects of studying unconventional nucleotides in general. Firstly, they are a part of life’s alphabet, and learning about different ‘letters’ used by different organisms shows us the boundaries of adaptations of living beings.
Since these modifications are possibly a part of the phage adaptation to constantly evolving bacterial viral defense mechanisms, learning about them contributes to our knowledge of the way viruses have adapted to avoid those defenses, and can instruct us in developing novel antivirals against eukaryotic viruses of concern. Additionally, previous research into bacterial anti-viral defense mechanisms have yielded important biotechnological molecular tools, such as CRISPR-Cas.
Since Naomivirus phages possess unconventional nucleotides, it is reasonable to assume that their DNA and RNA polymerases will be adapted to copy and transcribe from unconventional templates, so overexpressing and purifying these two enzymes can add to an arsenal of polymerases capable of producing unconventional DNA molecules, which would be resistant to existing host endonucleases and therefore be easier to transform into different bacterial hosts for cloning purposes, for example. Therefore, it is important to tap into this potential treasure trove of possible future biotechnological tools.
Finally, genomes of organisms that contain both conventional deoxynucleotides and unconventional ones, such as dUTP, can potentially teach us about the evolutionary origins of self-replicating molecules, especially if they contain nucleotides that bridge the gap between RNA and DNA.
References
Rihtman, B., Puxty, R. J., Hapeshi, A., Lee, Y. J., Zhan, Y., Michniewski, S., … & Chen, Y. (2021). A new family of globally distributed lytic roseophages with unusual deoxythymidine to deoxyuridine substitution. Current Biology.
- Main paper described in this feature
Takahashi, I. and Marmur, J., 1963. Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature, 197(4869), pp.794-795.
Kiljunen, S., Hakala, K., Pinta, E., Huttunen, S., Pluta, P., Gador, A., Lönnberg, H. and Skurnik, M., 2005. Yersiniophage ϕR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine. Microbiology, 151(12), pp.4093-4102.
Many thanks to Atif Khan for finding and summarizing this week’s phage news, jobs and community posts!