I’ve long been fascinated by REDCap, ever since I heard about it from Shawna many years back. Since then I’ve had the “pleasure” of building several REDCap projects of my own, for Phage Australia’s STAMP phage therapy clinical trial.
For our STAMP trial, we’re using REDCap in two ways: as a “screening and referral log” for evaluating and enrolling new cases, and as the clinical data collection tool, as an electronic case report form.
In this article, we’re introducing a short series called “REDCap Experiments” where I share some learnings from building on REDCap, and how we’re using REDCap for our STAMP trial.
What is REDCap?
REDCap stands for “Research Electronic Data Capture” and is kind of a Google Forms for collecting research study data from a range of fields, from social sciences and clinical studies.

Fig 1. On the left is REDCap, the electronic data capture tool. On the right is a GPT-drawn “redcap”, a goblin-like creature from Scottish folklore. They are not related.
The biggest difference between REDCap and Google Forms is that REDCap runs on a computer (a server) within an institution, while Forms runs on Google’s computers. For example, our server is administered by University of Sydney. By having a server at our own institute, we can safely and legally (adhering to “on premises” and “data sovereignty” laws) collect and store all kinds of sensitive, personally identifiable information (”PII”) about our patients.
This makes REDCap ideal for projects like clinical trials, where very sensitive information is collected from patients.
REDCap is built different
Software like Google Forms is usually backed by huge corporations. REDCap isn’t like that: originally a research project, the software is maintained by a small team of 5-10 employees at Vanderbilt University Medical Center and the REDcap Consortium, and is free for institutions. Despite its grassroots history and its tiny team, it’s used across almost 7,000 institutions in 155 countries, and has 35k citations!
Unlike Google Forms, REDCap is research software. This means it lacks the depth of documentation and support that corporate software has (Google Forms has a huge section for software developers to extend it with features).
Learning REDCap means finding PDFs written by university librarians of various levels of quality. Many of these PDFs are many years old, incomplete or incorrect. Very few are written by those who have successfully built large projects on REDCap.
All this makes learning REDCap incredibly difficult.
For our STAMP project, we’ve had to learn REDCap the hard way by building four iterations over two years, before finding a way that works for us. And I’m still not sure if we’re doing it the “correct” way!
Building projects on REDCap
The best way to learn REDCap is to build projects on REDCap.
The rest of this article will explore some ideas I’ve built on REDCap, around a fictional “Ice Cap” ice cream ordering system. If you have access to REDCap (through your institution) and you’d like to follow along, I’ll be sharing links and data dictionaries along the way!
The rest of the article will get fairly technical. If you’re completely new to REDCap and want to know where to start, there’s some intro summary and training videos modules under the training resources page. Check back once you’ve had a go at the Online Designer and the Data Dictionary!
Before you start…
…put aside everything you know about forms, databases and web frameworks. REDCap doesn’t work the way you expect it to work. In REDCap, there’s no components or javascript or databases. You have to understand the REDCap mental model, and it affects how you start designing projects.
When we started building out our STAMP protocol on REDCap, we wanted lots of conditional questions: how many pathogens are we targeting? For each pathogen, how many susceptible phages are available? For each pathogen/phage/antibiotics combination, what are the details?
Building this in Javascript is relatively straightforward: you show the pathogen/phage/antibiotic form for each combination, with a combination of loops and if statements. In REDCap, this is less straightforward.
There’s two ways to accomplish this task in REDCap. The first, preferred, idiomatic REDCap way is to use *repeating instruments* where you have separate instruments (forms) for each item you want to collect multiple times, like isolates, phages, and antibiotics. This can get tedious though, as you have to fill out many forms, and often you’ll have to duplicate information across those forms.
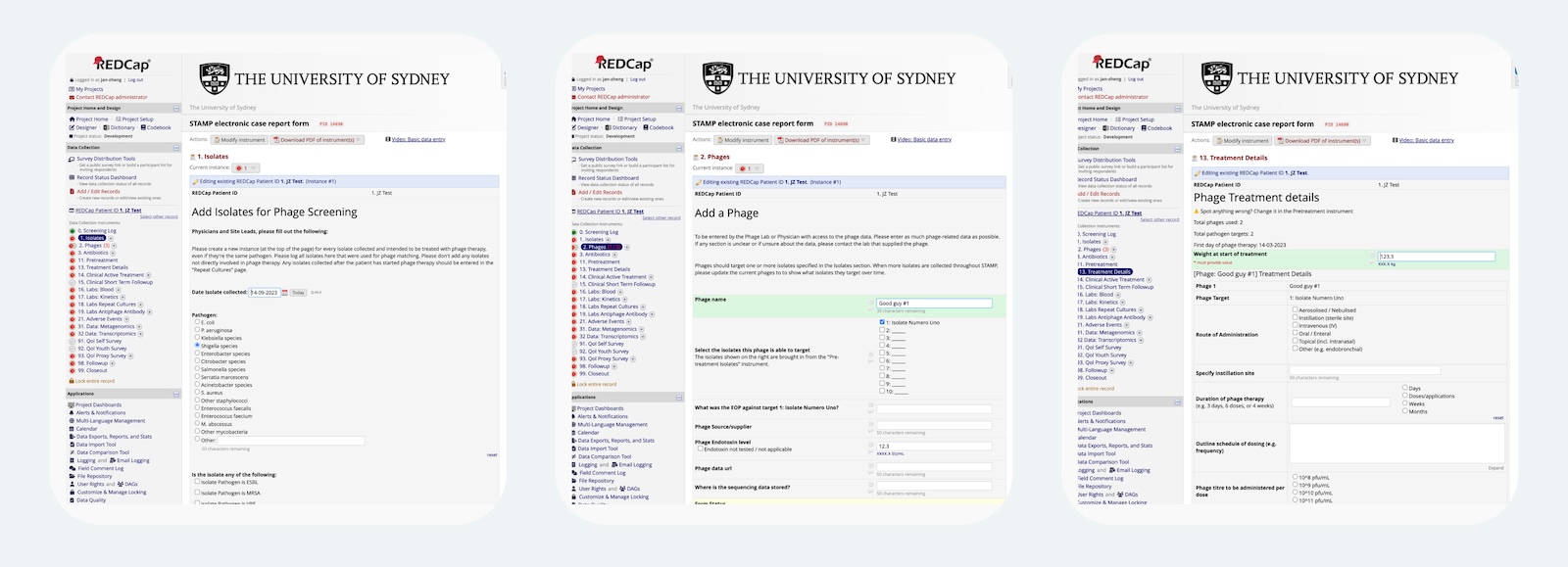
Fig 2. For STAMP, our Isolate and Phage instruments collect information about each item, and the Treatment Details instrument lets users enter details about each isolate x phage combination. Full image
In the first two panels, we collect isolates and phages with repeating instruments. In the third panel, we ask about every combination of phages and pathogens in a single instrument. Putting all these questions on a single panel makes the information easier to enter, view, and understand. Combining these questions in a single form, in a dynamic manner isn’t built into REDCap behavior… so how does it work?
Experimenting with permutations in REDCap
If you’ve worked with REDCap, you’ve probably wanted to use loops or permutations. For example, “please enter data for every combination of X, Y, and Z” or “how many ice creams would you like? What would you like on each order?”
This kind of question structure is surprisingly hard to build, as REDCap doesn’t support scripting, so you can’t use loops or if statements. But if you combine the existing logic tools with permutations, you can accomplish something that looks and feels dynamic.
Before we continue, fill out this imaginary Ice Cap Ice Cream shop form: https://redcap.sydney.edu.au/surveys/?s=DLKJJTXCA8FCCD3M
Ice Cap is a toy example I created as a way to experiment with REDCap, without collecting or messing with sensitive clinical data — it’s a simplification and teaching tool that encapsulates some of the things we’ve been doing for the STAMP phage therapy clinical trial.
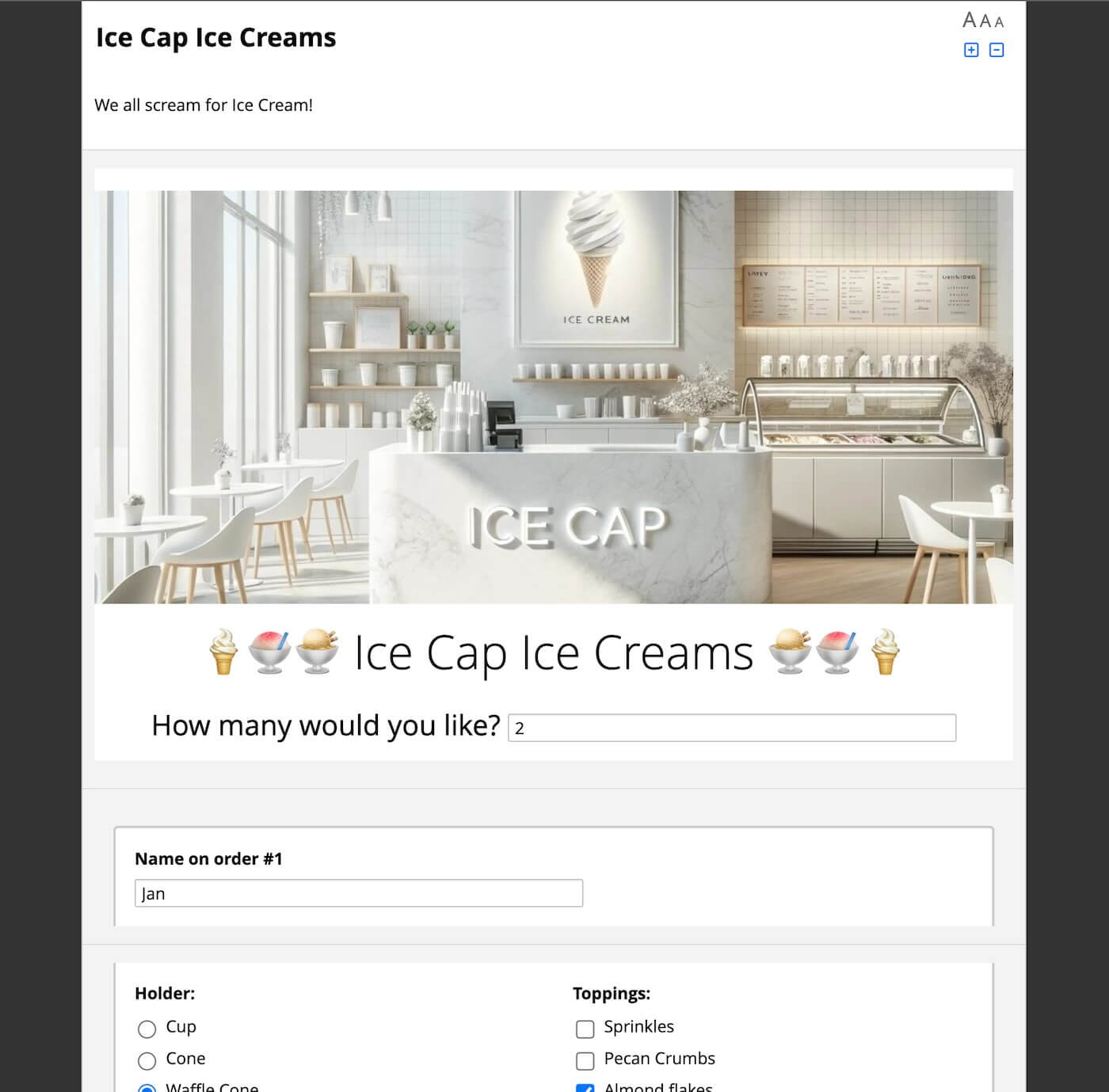
Fig 3. Ice Cap Ice Creams is an experiment in building loops and permutations. It dynamically creates a number of ice cream orders and flavors based on how the number of orders and scoops are requested. Fill it out here!
In this example, we can ask users how many ice creams they would like. After they say a number, we ask them what container and toppings they’d like, for each ice cream order. We then ask how many scoops a user wants, and display the Scoop form for each flavor dynamically. If the user requests 9 flavors, we show 9 different fields.
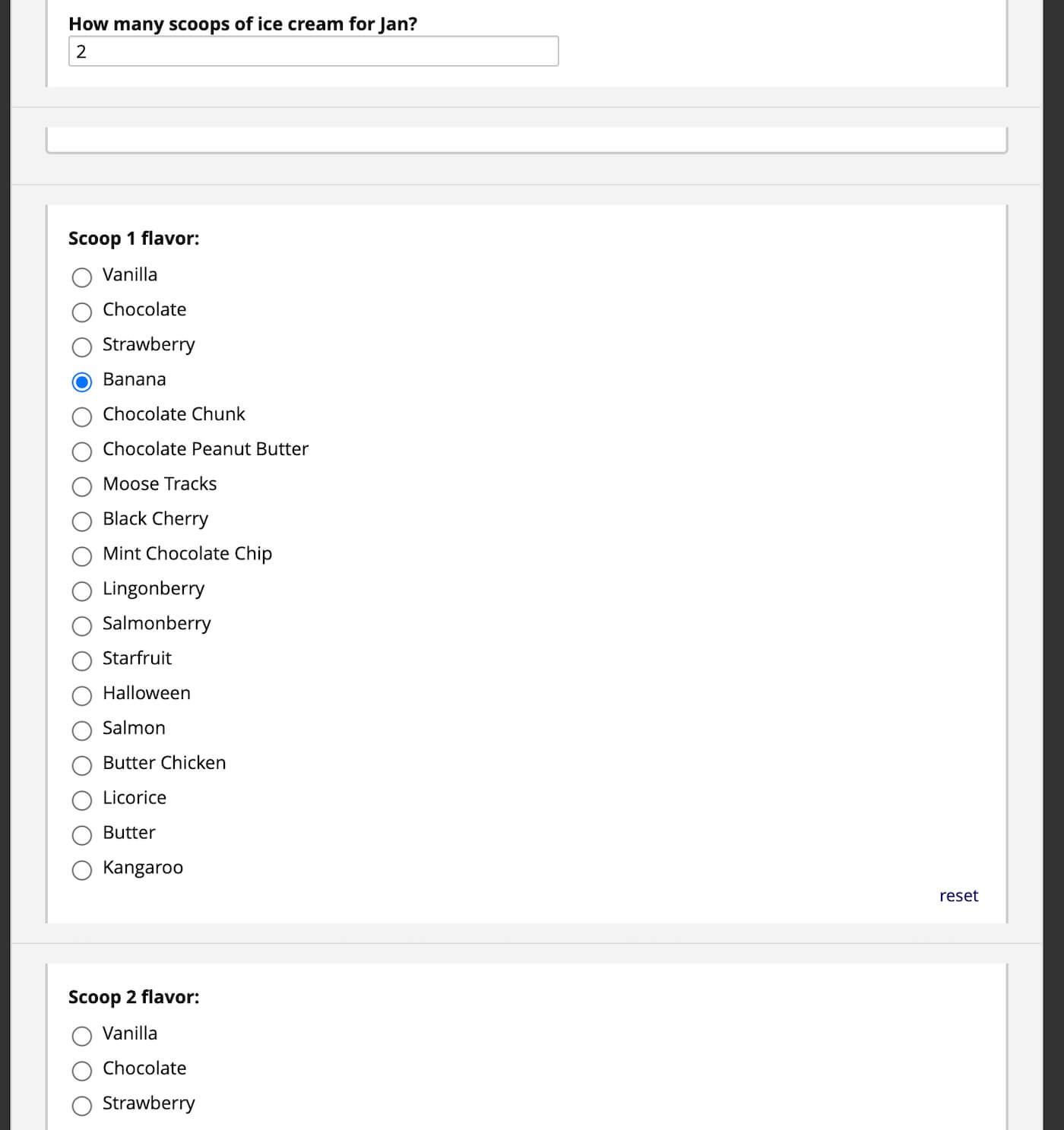
Fig 4. We can ask for a number of ice cream flavors for each scoop, which is assigned to each ice cream order.
How do permutations work?
Under the hood, setting up permutations is fairly simple: we create a separate form question for every possible ordered scenario of ice cream orders, scoops, and flavors. For example, if you have three (3) questions, and each question asks to choose between two (2) options, you’ll have 2 × 2 × 2, or eight different questions.
The Ice Cap example that you just filled out, though fairly simple, has a total of 129 different questions — every question needs to be accounted for. This also means that we need to limit the number of orders and scoops a person could order. For example, I limit the number of orders to 5, and the number of scoops to 9, though these are arbitrary choices. The higher you set the limits, the more form fields will need to be created.
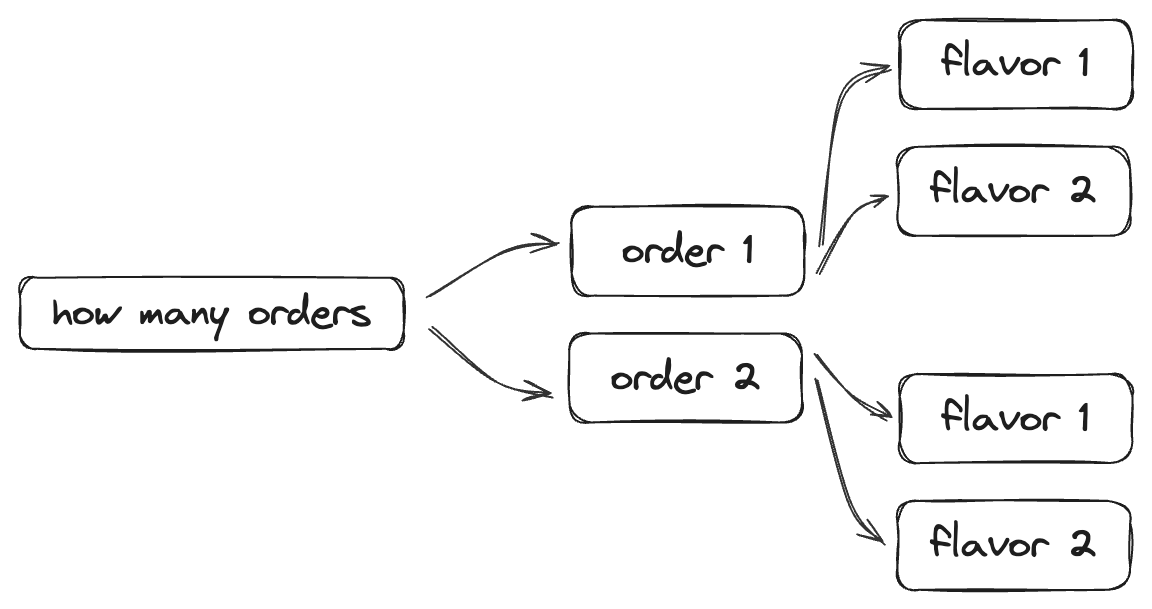
Fig 5. When dealing with permutations, we need to create every possible question that could be asked. In this simple scenario, if we limited the number of orders and flavors to two, we would create 7 different form fields in REDCap.
Once all the questions are created, we use branching logic to show or hide each field based on the choices and numbers users enter. For example, if a user requests 3 orders, we hide all the fields that are created for orders 4 and above.
The following is a series of progressions of REDCap instruments that shows how such a system can be reasoned about and built incrementally.
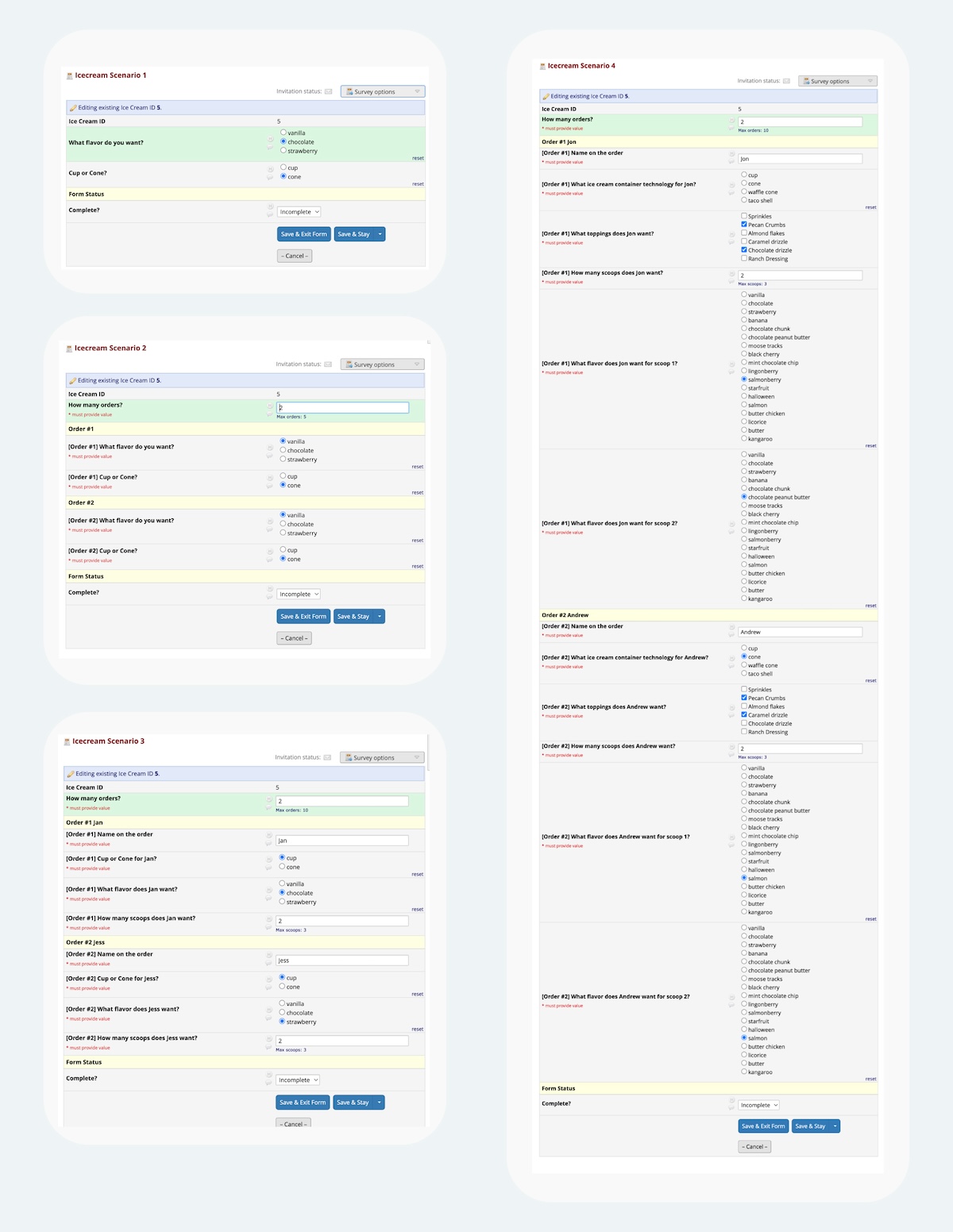
Fig 6. During the development process to figure out REDCap’s capabilities, I’d created these four different increasingly complex Ice Cap ordering scenarios. The first scenario has no permutations, and only uses two fields. The second scenario displays order forms up to five orders, and uses 11 fields. The third scenario is similar to the second one but has a limit of 10 orders; this uses 41 fields. The fourth scenario shows how you can “nest” permutations, by first asking for x number of orders, then for each order, ask y number of scoops; this uses 71 fields. Functionally, scenario 4 is similar to the final example, but with fewer choices and without the fancy bells and whistles. Full image
If you’re thinking about using combinations or permutations in your own REDCap, play with each of the following scenarios, and look at how the data dictionary is constructed:
Scenario 1: https://redcap.sydney.edu.au/surveys/?s=NWXX9MoEfVkcxDg3
Scenario 2: https://redcap.sydney.edu.au/surveys/?s=cpE6cZ2RbgS8jWwv
Scenario 3: https://redcap.sydney.edu.au/surveys/?s=8F3NsZ6AfszDLnL7
Scenario 4: https://redcap.sydney.edu.au/surveys/?s=XbfJGXXRaNHYrE98
Scenario 5: https://redcap.sydney.edu.au/surveys/?s=DLKJJTXCA8FCCD3M
Data Dictionaries: https://docs.google.com/spreadsheets/d/1L1ePt2wiUQwgCSj5e7Z_-CNNl4iomXbihrhyvCOLRbw/edit?usp=sharing
Variable names are important
For the Scenario 5 data dictionary, I’ve named the variables as s5_order1_ ... to help me keep track of different variations. You’ll see that for each scenario, order, and scoop, some variables act purely as descriptions, while others act as fields for data collection.
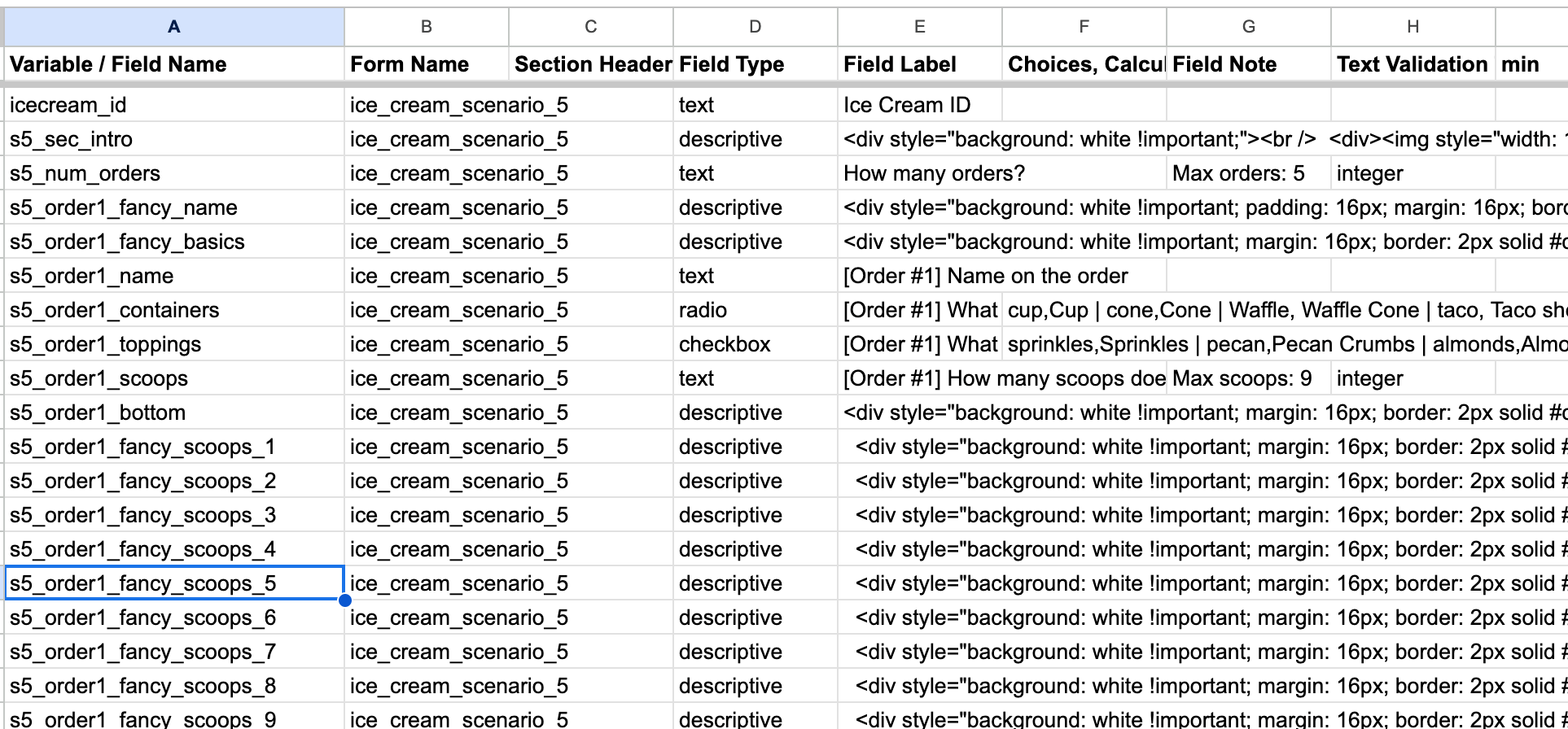
Fig 7. Notice how the variable names use a combination of names, numbers, underscores, and shorthands to make it quick to understand combinations / permutations, and what each field is supposed to do.
Create permutations one layer at a time
For our STAMP trial, we combine nested permutations with repeating instruments, and the number of combinations of fields can get complicated — especially when you’ve found a bug or a problem. The way we built our STAMP REDCap is similar to how I broke down the five scenarios of the ice cream store: once you’ve nailed down one layer of questions — for example choosing between scoop/cone type and flavor — you can then continue to the next layer of questions, like number of orders. I highly suggest creating each layer of questions separately, then combining them.
Fixing bugs and correcting typos is hard
Using Google Sheets to manage a data dictionary with permutations gets tricky. The “Find and Replace” functionality is your best friend, because many texts are repeated across each variation. “Find and Replace” is great for correcting simple typos or logic problems, as you can quickly replace multiple rows in one go.
Formulas are your friend
If you want to get fancier, you can assemble permutations and variables using a combination of formulas like arrayformula(flatten(transpose())) with sequence(). If you deal with a lot of repetition, you can create those definitions in another table, and use vlookup() ,substitute() , and mid() to specify exactly what texts to replace. This can get tricky and probably deserves its own post, so I’ll reserve this for a future article.
ChatGPT is your friend
If you get stuck, especially with formulas, it’s ok to ask ChatGPT for assistance. I found it great for helping with explaining and even generating formulas for me.
It can even generate permutations of the data dictionary for you, without you having to resort formulas! Just make sure you double check the answers as it’s prone to making mistakes. Also, make sure to use GPT-4 if you’re generating either formulas or permutations.
Final thoughts
For phage clinical trials, being able to create forms with combinations and permutations can be very powerful, as the data we collect depends on many variations and combinations. Permutations give us the flexibility to ask specific questions about those combinations.
Even with the help of formulas and ChatGPT, large projects with complex permutations can quickly get out of hand. For the next REDCap Experiments article, I’ll write about how we design and build our data dictionaries, with a combination of Google Sheets, formulas, ChatGPT, and a specialized programming language for schema definitions, called CUE lang. (All of the Ice Cap experiments were created in CUE!)
This article veers a bit away from our regular phage articles, but I think having a good grasp of tools like REDCap is absolutely crucial for the future success of phage therapy clinical trials.
If you have any questions about this article, (or general REDCap questions) you can email me — I’ll be happy to answer them: [email protected]
Best,
Jan
References
Ice Cap Data Dictionary: https://docs.google.com/spreadsheets/d/1L1ePt2wiUQwgCSj5e7Z_-CNNl4iomXbihrhyvCOLRbw/edit?usp=sharing
Scenario 1: https://redcap.sydney.edu.au/surveys/?s=NWXX9MoEfVkcxDg3
Scenario 2: https://redcap.sydney.edu.au/surveys/?s=cpE6cZ2RbgS8jWwv
Scenario 3: https://redcap.sydney.edu.au/surveys/?s=8F3NsZ6AfszDLnL7
Scenario 4: https://redcap.sydney.edu.au/surveys/?s=XbfJGXXRaNHYrE98
Scenario 5: https://redcap.sydney.edu.au/surveys/?s=DLKJJTXCA8FCCD3M
Harris, P. A., Taylor, R., Thielke, R., Payne, J., Gonzalez, N., & Conde, J. G. (2009). Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. Journal of biomedical informatics, 42(2), 377–381. https://doi.org/10.1016/j.jbi.2008.08.010













