Ask people on the street to name the most common forms of life on Earth, and you’re sure to get a bevy of answers. Some, recalling those riveting high school lectures on photosynthesis, might say plants. Others, knowing that Earth is largely a water world, might say fish. Those fretting about overpopulation might say people.
A few may actually know that most life is invisible to our eyes, and name bacteria or even archaea as the most numerous. Yet even they would be wrong. Phages — the viruses that prey on bacteria and archaea — are actually the most numerous form of life. Scientists estimate that there are 10^31 phages out there, or roughly one trillion for every grain of sand on the planet. That’s a lot of life to leave unacknowledged in the streets, and unexplored in the lab.
So much, and yet so little
It has been more than a century since these miniscule viruses were discovered. But beyond learning the broad trends of phages — and studying a handful of “model phages” in detail — the most numerous form of life remains the least understood. And the history of science has shown us that understanding life often leads to great leaps in our own lives.
Phages do much that piques our interest. We know their activity shapes the bacterial and archaeal composition of microbes in every ecosystem, from soil to our stomachs. Phages are also agents of horizontal gene transfer, which spreads antimicrobial resistance genes. And though they cannot infect us, phage activity impacts our health. But nearly all information about the phage realm – including those fine details that are critical to understanding phages and combating the spread of antimicrobial resistance – remains buried in the sand. We need data spanning the gamut of phage biology including species interactions, abundance, diversity and complexity, to forge new advances in science and medicine.
Phase Genomics is pioneering an endeavor to end this wandering in the desert by developing the world’s largest repository of phage interactomes — interactions between phages and their hosts in natural ecosystems — aimed to enable new, phage-based therapeutics against infections and diseases as well as new methods for microbial surveillance. This effort, which recently received $5.5 million in support from the Bill and Melinda Gates Foundation and the National Institute of Allergy and Infectious Diseases, will combine Phase Genomics’ ability to map host-virus and cell-cell interactions within microbial communities with AI-driven predictive engines to tap the therapeutic potential of the phage realm.
Proximity promises
Our approach recognizes the reality that whole genome sequencing and assembly alone cannot address historic under-investments in phage research. The problem is not just a lack of knowledge about the diversity of phages out there. Scientists need rich information about interactions between phages and their microbial hosts in natural settings, however normal DNA sequencing tools cannot detect these interactions since virus and bacterial genomes are usually disconnected pieces of DNA.
One of the two main tools in this new effort, Phase Genomics’ ProxiMeta platform, is tailor-made to capture this knowledge. ProxiMeta is a platform for metagenomics that uses a unique DNA sequencing technology that can assemble and map virus genomes and interactomes in microbial ecosystems without the need for culturing in a laboratory. A paper published earlier this year in Nature Biotechnology reported that ProxiMeta could identify hundreds of novel host-virus interactions in just a single microbial sample.
Using ProxiMeta, Phase Genomics will construct comprehensive phage-bacterial interactomes for more than a thousand clinical, agricultural, environmental, and wastewater samples from around the world. This approach will not only identify novel phages, but also reveal their preferred host species and strains, as well as the mobile genetic elements and antimicrobial resistance genes that they help spread. The second arm of this endeavor — predictive engines powered by AI — will mine interactomes to identify phages of potential therapeutic value.
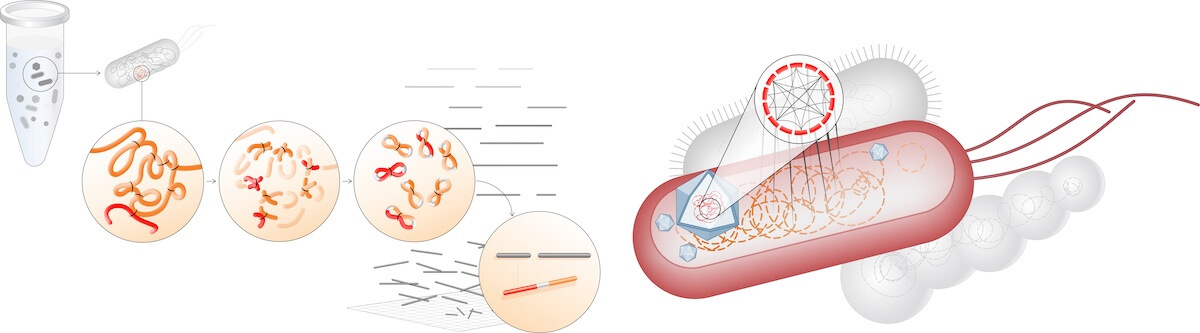
Overview of proximity-guided metagenome deconvolution and host attribution. Metagenomic samples are comprised of complex populations of archaea, bacteria, and fungi. In addition to chromosomal DNA, cells contain genetic material such as plasmids, viruses or bacteriophages, transposons, integrons, and other mobile genetic elements. Proximity ligation data provides an additional layer of information that is used to reconstruct more high-quality bacterial and viral genomes than traditional binning approaches, enabling accurate viral-host attribution. Image courtesy of Phase Genomics, Inc.
So little, and yet so much
Research indicates there’s a lot of phage potential to mine. The spread of antibiotic resistance has led some scientists to revisit phage therapy as a method to treat infections — an option abandoned in its infancy after antibiotics stole the spotlight nearly a century ago. Inflammatory conditions like Crohn’s Disease and ulcerative colitis show altered phage distribution in the gastrointestinal tract, pointing to phage tinkering as a potential treatment. In our efforts, AI predictive engines will seek out therapeutic phages against these types of conditions. But with so few phages known among that 10^31, there are likely a lot of surprises out there, and further potential beyond these initial goals.
Time is not on our side. By mid-century, antibiotic-resistant infections may surpass cancer as a cause of death worldwide, and add $1.2 trillion to global health expenditures. But urgency should not mean a mad dash to dig wildly in the sand. We need maps to the phage realm. And now, fortunately, we can build them, allowing a targeted search through the desert that at the same time will illuminate one of biology’s biggest blind spots.








