About two years ago, I completed an internship at Dr Paul Jaschke’s lab at Macquarie University (Sydney, Australia). I was trying to evolve a decompressed phiX174 bacteriophage created by my supervisor several years ago at Stanford University. One of the tasks was to measure plaques of the evolved phage. I looked online for the tools to ease my work and found a couple that seemed suitable, but I struggled with their installation. After installation, I realised that they required some additional settings that weren’t easy to understand, and they could not process plaques with bull’s eye morphology (plaques with a clear centre and one or several outer turbid concentric rings) either. So, I gave up, found a ruler in a drawer of one of the lab benches and measured the plaques manually. The work was finally done.
This year I needed to measure plaques of different sizes and morphology for my own project; some of the plaques were extremely small, and it would have taken forever to assess them manually. So, this time, I’ve decided to refresh my software engineering skills and created a plaque size measurement tool called Plaque Size Tool based on the OpenCV Python library. It was published in Virology (Elsevier) in 2021, and its files are available on GitHub.
Installation
Because of my previous frustrations with installing various tools, I’ve tried to make the installation as easy as possible. It requires pre-installed Python 3, but all other packages are installed automatically, so there is no need to install them manually one by one. I’ve also added screenshots to the manual so you can follow along.
Validation
To check the tool, I first tested it on 499 plaques formed by different variants of phiX174 phage on E. coli C and found that the results did not significantly differ from the manual measurement in Fiji (where you need to circle each plaque manually). After that, I looked for other images of plates with phage plaques and found an extensive collection in the Actinobacteriophage database. Processed images of various formats and quality can be found in the Supplementary File of our paper.
During peer review of the Plaque Size Tool, one of the reviewer’s questions was whether the tool could handle turbid plaques as they do not have clear edges due to incomplete lysis. With the great help of the Phage Directory community, we were able to obtain additional plate images with turbid plaques and measure their plaque size. Overall, the tool can measure between 70 and 100% plaques on a plate for plaque sizes greater than 1.5 mm and 51-91% for plaques less than 1.5 mm. Although not designed to measure titres, if all plaques are distributed on the plate without forming overlaps, it is possible to get the total number of plaques on a plate. The tool can measure sizes of plaques of quite different morphology: clear, bull’s eye, turbid, and plaques with clear centres and turbid edges.
Additional options
The tool can measure the plaque size (diameter, area) either in pixels or in mm. To get the value in mm, a plate size in mm needs to be specified in the option -p (for example, -p 90 means that the plate on the image is 90 mm in diameter).
The PST can process individual images (-i option) and bulk if a directory with images is specified (-d option). If you’re processing photos with small plaques (less than 1.5 mm), another option could be used: -small. In addition to the output image with valid plaques outlined with green colour and ID with diameter specified for each plaque, a CSV file is also formed containing all plaque IDs with corresponding Area and Diameter values both in pixels and mm if plate size is specified.
The example of the images processed with the PST shows two plates: one with small plaques (top) and another with average plaques (bottom):
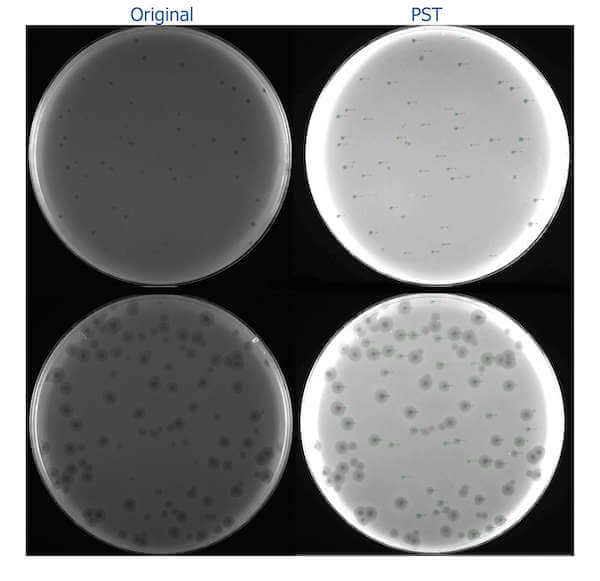
As testing was done on a large but still limited sample of images, it is possible that the tool might not process particular images very well (for example, the images are too bright or dark or have noise). In that case, I would appreciate it if you could send me an email ([email protected]) or create an issue on GitHub to fix a particular issue and ease your work of phage size measurement. The tool works under all OSes supporting Python 3, is open source (under Apache License 2.0), can be used in any automatic pipelines and modified by users for other purposes.
—
Many thanks to Atif Khan and Stephanie Lynch for finding, summarizing and editing this week’s phage news, jobs and community posts! And thanks to Lizzie Richardson for editing the feature article!