Dear Phage community,
Since this is my first post to the community, I thought I’d introduce myself.
Hi, I’m Aaryan! Right now, I’m working with the Bollyky Lab at Stanford University, specifically on phages to target resistant strains of P. aeruginosa. Beyond that, I love building tools to support the phage community.
In this article, I’m announcing the launch of PhageBase (www.phagebase.com) — the world’s largest atlas of bacteriophage images.
But before I explore how PhageBase can accelerate and promote your research, let’s think about why this matters at all.
How It Began
PhageBase was born from an interaction I had with my former mentor, Arne Echterhof. At the time, Arne just received a new phage called Terra from one of our collaborators. All he wanted was to find its dimensions so he could embed it in a hydrogel with the right pore size. But it took him nearly a week of tortured searching to find what he was looking for.
Why did Arne have to spend so much time away from his research scouring the internet for public data? Why wasn’t such basic information about phages available in one place?
Later that weekend, I put together a janky HTML site called PhageBase to host data on our lab’s most common phages. Soon enough, though, we realized our lab wasn’t the only group using the platform. So we continued to polish the site, expand our dataset, and iterate on feedback. And now we’re here.
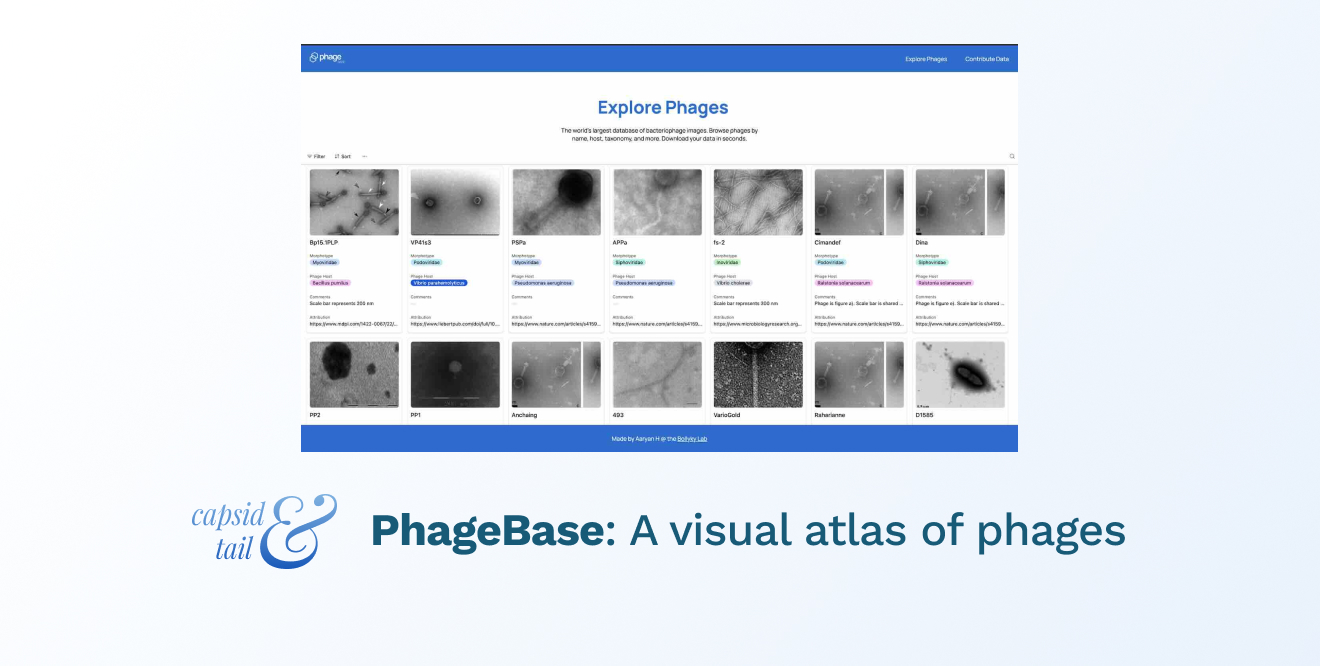
The user interface for PhageBase
Where We’re At
To our knowledge, PhageBase is now the largest repository of its kind, with structural and micrographic data on over 1000 phages spanning 150+ bacterial hosts. Here’s how you can use it to accelerate your research:
Exploring Phages
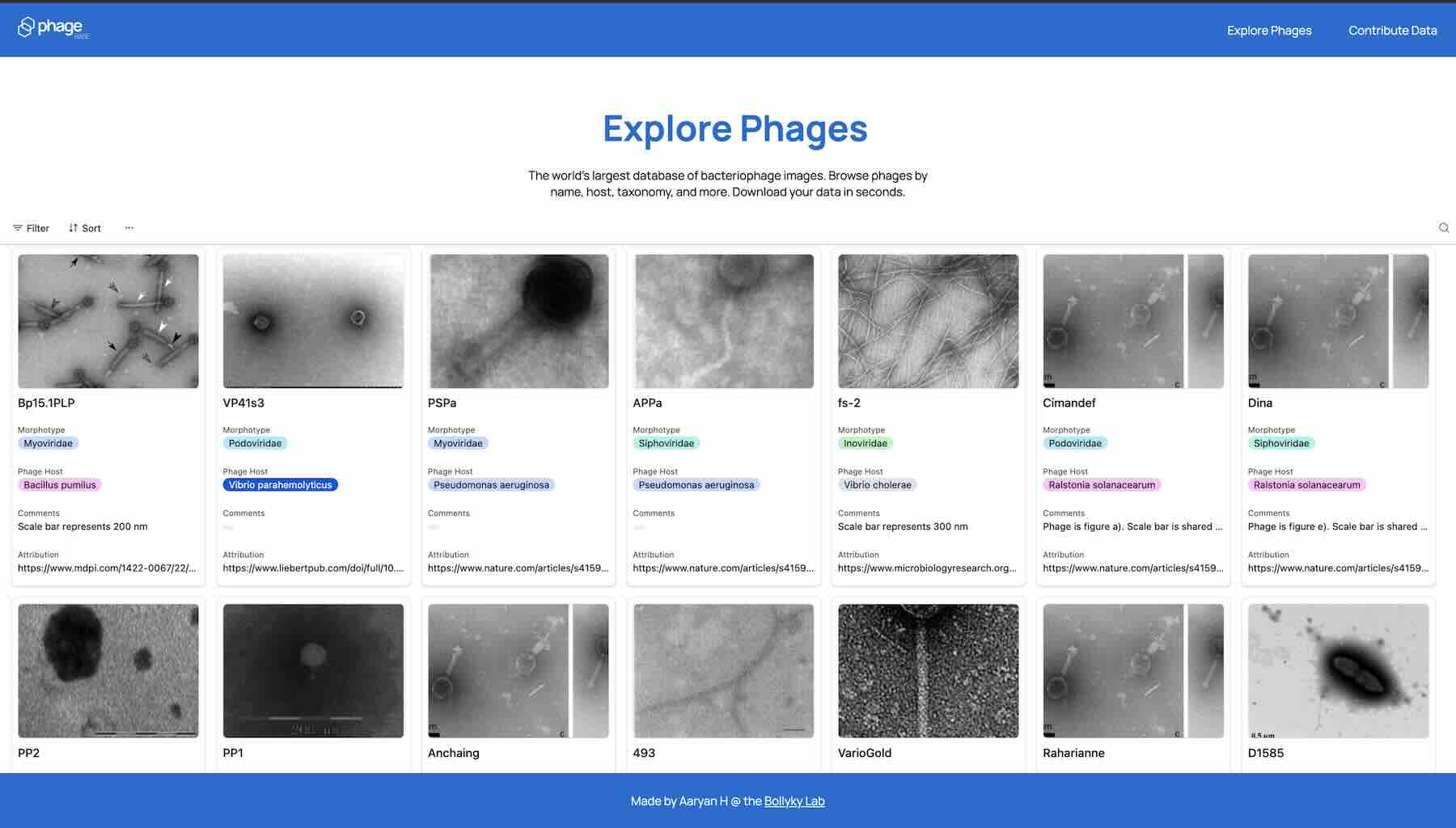
PhageBase helps you find phages faster than Google Scholar. And since the platform focuses exclusively on phages, it lets you filter by name, host, and morphology.
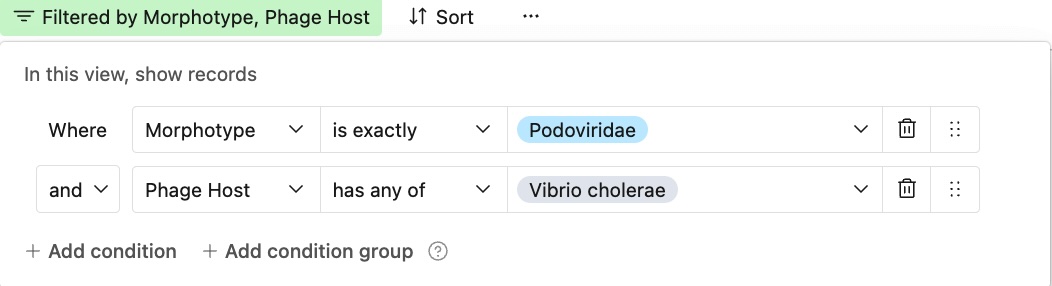
The “Filter” and “Sort” tools are available at the top left of the database window
Example: To find all the podophages that infect V. cholera, just set the “Host” to V. Cholera and the “Morphotype” property to podovirus. Each entry contains a detailed register of information about the phage, all of which were inputted and cross-checked by human reviewers. To learn more, just click the link in the “Attribution” section to read the paper where the phage was originally described.
In short, PhageBase lets you find data on virtually any phage instantly. That means less time scouring obscure papers for tidbits of data. Which leaves more time to run your experiments, get published, and test out new ideas.
Contributing Phages
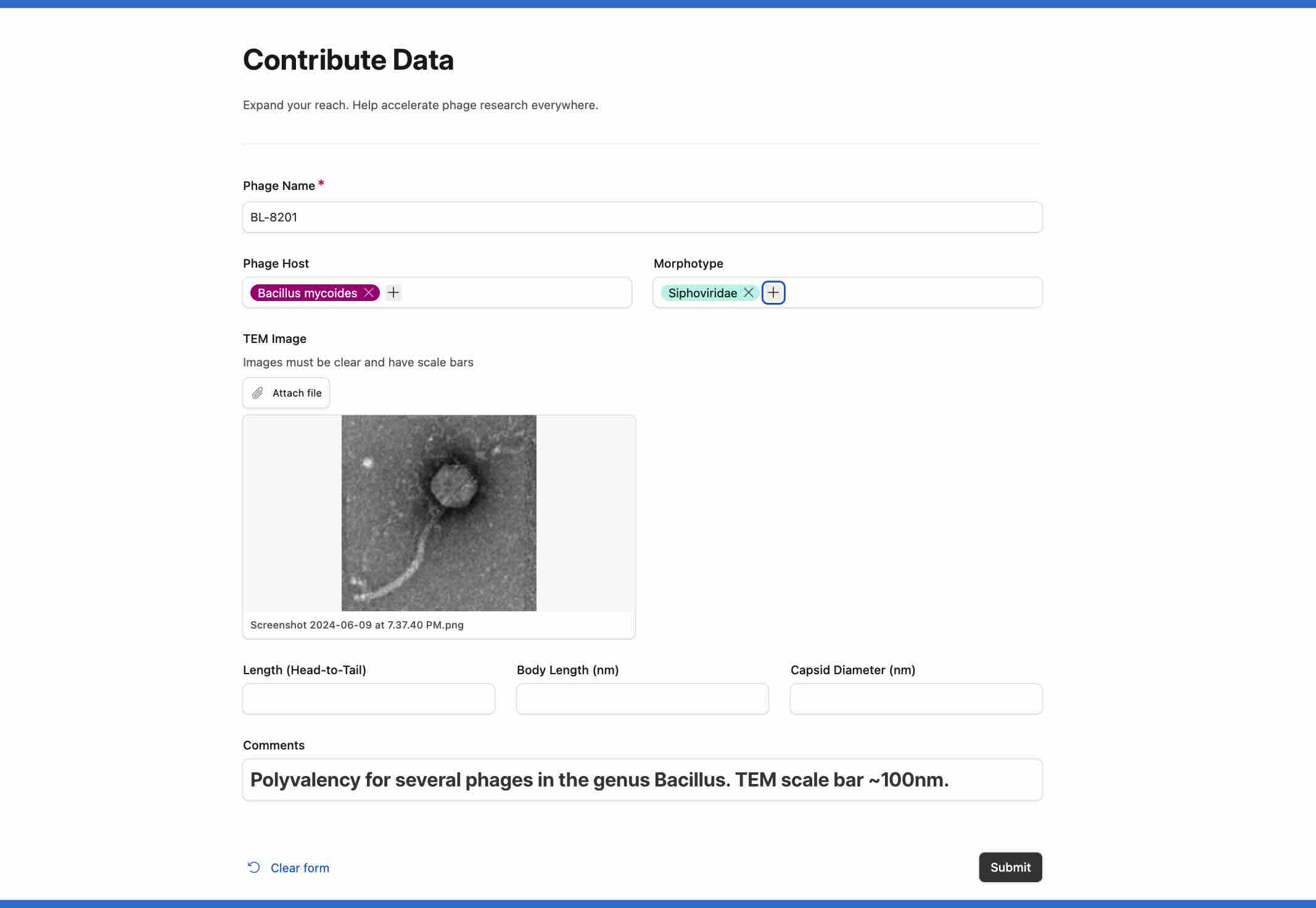
PhageBase was designed to be an open exchange. We want it to grow into a thriving community, where people can learn with and from each other.
Example: If your lab just isolated a new phage, use our Contribute Data form to make it visible to researchers around the world. If your lab just published a paper, use the same form to spread the word and get more citations. Aside from advancing your career, this is a simple practice to give back to the phage community.
Finding Collaborators
From speaking with our fellow researchers, we noticed that it was still tough for our community to connect. We wanted to change that.
Example: Did you have a question about one of the papers on PhageBase? Or, are you trying to buy a phage that just isn’t available online? What if you just want to have a conversation with a research group halfway across the world? Just open up the paper listed on PhageBase and email the corresponding author.

Next Steps
Of course, these features represent a fraction of what PhageBase could be. We’re still adding phages to our repository and brewing up new ideas every day.
By the end of the year, we want PhageBase to reach its first 10,000 phages. We also want to grow the list of parameters offered for our entries (i.e. burst size, genome length, etc.) and extend the platform into an API. And, of course, we want to launch systems that make it easier for our community to collaborate and build on each other’s work. In other words, there’s a lot left for this platform to give.
Where We’re Headed
At one point, for personal computing to mature, we had to transition from clunky, home-built transistors, to precision-engineered chips fabricated on assembly lines. For synthetic biology to mature, we first had to systemize the art of gene expression, sequencing, and purification.
History shows us that most of our globally relevant technologies began in the labs and garages of a handful of passionate tinkerers. But for these tools to develop from pet projects into industries, all of them first had to be organized and developed. Standards had to be set. Infrastructure had to be built. Data had to be centralized.
In the grand scheme of phage therapy, we find ourselves at this exact turning point. And as the founding tinkerers of this field, we get the chance to write the rules that define its future.
How You Can Get Involved
We want PhageBase to become a hub that enables this future. If our mission resonates with you, there are three main ways you can join our journey.
-
We need more people talking about PhageBase. If you share PhageBase on X and The Phage Directory (and send me screenshots of both at [email protected]), you’ll get a chance to win one of three T4-phage plushies.
-
We’re putting together an informal board of directors to steer PhageBase over the long term. If you either run a phage lab, help lead people at a phage startup, or know someone who fits this description and might be interested, email me at [email protected].
-
Right now, we’re getting more phage contributions than we can review on our own. If you’re interested in volunteering with us and gaining meaningful experience with phage research…again, email me! (While we think this opportunity is probably most valuable to high-schoolers and university students, we welcome anyone who cares about phage research).
Of course, I’m also curious to hear what you think about the platform. What are your initial reactions? Where do you think we can improve?
Time after time, this community has surprised me with its drive and generosity. I find myself counting on you again. Let’s rely on each other to advance our research. Let’s make order from the chaos that surrounds us. Together, let’s make phage research as exciting and promising as we all know it can be.