In this week’s feature article, we’re highlighting a paper recently published by Shelley Gibson and colleagues at Baylor College of Medicine and the Michael E. DeBakey VA Medical Center in Houston, Texas.
Through academic-clinical collaboration, they’ve constructed and characterized phage libraries against three pathogens, and have made them available for phage therapy.
This effort serves as a great example of systematic phage characterization, data collection and presentation, which is increasingly needed in the phage field.
Main source:
Gibson, S. B., Green, S. I., Gu Liu, C., Salazar, K. C., Clark, J. R., … & Ramig, R. F. (2019). Constructing and Characterizing Bacteriophage Libraries for Phage Therapy of Human Infections. Frontiers in Microbiology, 10, 2537. doi: 10.3389/fmicb.2019.02537
Which pathogens do the phage libraries target?
- Escherichia coli (ExPEC)
- Enterobacter cloacae
- Pseudomonas aeruginosa
How were the phages characterized?
- Isolation source and host
- Host range (efficacy against a panel of clinical strains)
- Basic infection parameters (burst size and adsorption rates)
- Genome sequence analysis (looking for lysogenic, antibiotic resistance or toxin genes)
Which strains were used to determine host range?
The team collected clinical isolates of all three pathogens from the clinical microbiology lab at the Houston Veterans Administration Hospital and Baylor St. Luke’s Hospital. Isolates mostly came from urine and blood of patients with urinary tract infections (UTIs) (including spinal cord injury patients with urinary catheters). Others came from left ventricular assist device (LVAD) infections.
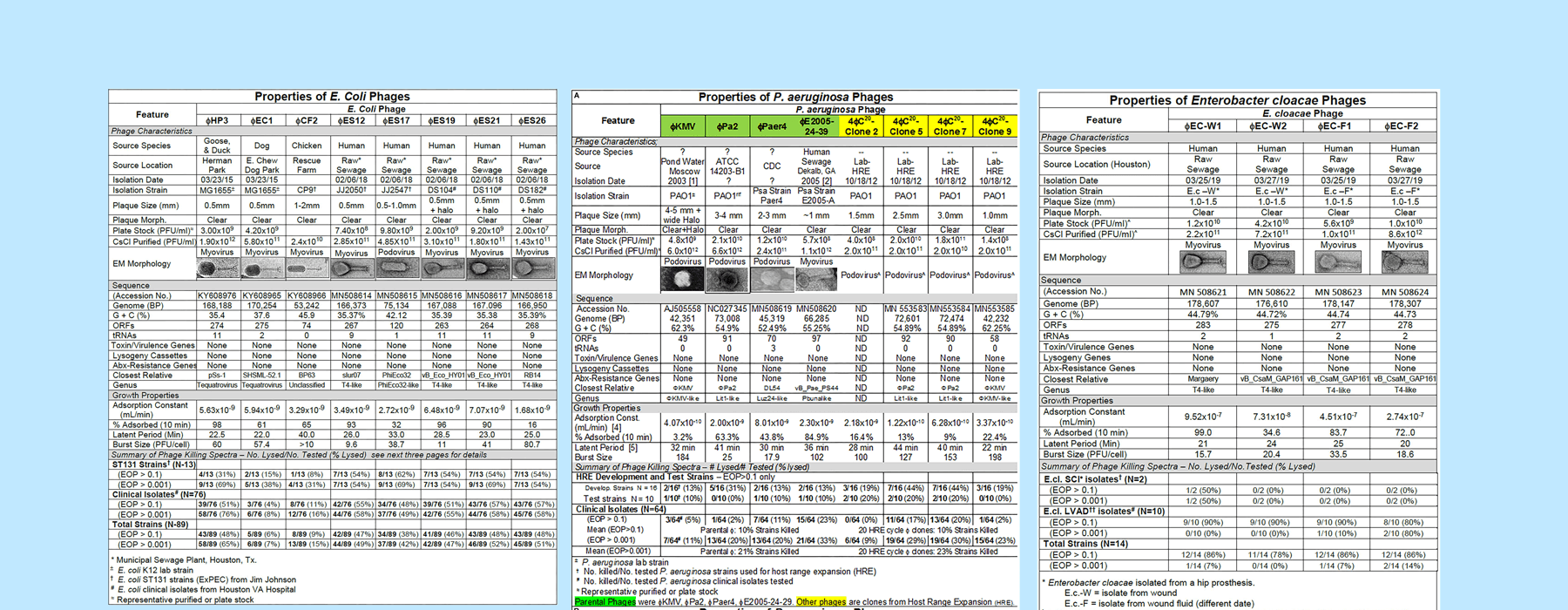
E. coli phages: coverage of most clinical isolates through cocktails
The E. coli phages the team had in the lab didn’t plaque on ExPEC ST131, their strain type of interest, so they isolated eight phages from environmental samples (avian and canine species are known to harbor ExPEC ST131; they found phages in goose, duck, dog, and chicken feces from parks and farms, as well as from a sewage plant).
A cocktail of three of their eight phages could kill 92% of their 76 clinical isolates. No single phage killed more than 50-55% of the strains.
All phages got to high titres and had varied biological characteristics.
E. cloacae phages: coverage of most clinical isolates without need for cocktails
Four E. cloacae phages were isolated from sewage samples using a phage therapy candidate’s isolate as a host.
They tested the phages on clinical isolates from LVAD infections and UTIs. 70% of the strains were killed by all individual phages (cocktails didn’t improve this).
All phages were T4-like and similar to one another.
P. aeruginosa phages: coverage of most clinical strains through host range expansion and cocktails
Four previously described phages (ϕKMV, ϕPA2, ϕPaer4, and ϕE2005-24-39) were shown only to lyse 2-23% of the team’s 64 clinical strains. To broaden their host range, they subjected these phages to a host range expansion technique (also known as the Appelmans protocol, which is used by Eastern European phage researchers).
They tested the host range-expanded phages on their 64 P. aeruginosa clinical isolates, and found that a three-phage cocktail of the new phages could lyse 81% of them. However, many phages killed at too low an efficiency to be useful for therapy, suggesting room for improvement of this library.
The host range-expanded phages had similar growth properties to those of their parental phages.
How were phages analyzed genetically?
Genome sequencing was done for all phages (analysis was done in part through PATRIC, a free comprehensive genome analysis service). Genomes were scanned for virulence factors and antibiotic resistance genes. They scanned against several databases, including:
They also scanned the genomes for evidence of a lysogenic lifestyle (integrases, attachment sites) using:
Did they find any detrimental genes?
Most phages appeared devoid of toxic and lysogenic elements and thus appear safe for phage therapy. One E. coli phage contained a suspected integrase gene, though it could not be induced by mitomycin C.
Next steps
Working with local clinics to make phages available for more patients
This group plans to work with their local Veterans Affairs Hospital to have phages available for testing on each patient strain that comes in, during the time it’s being tested for antibiotic resistance. If phages are found active against the isolate, it will be the physician’s choice to use phages, antibiotics or both.
They also plan to keep collecting strains from the urine of all spinal cord injury patients that come into their hospital, and ultimately to create phage libraries that can cover any infection (with any pathogen) that comes in. They’re doing the same at the Baylor-St. Luke’s hospital for all bacterial strains causing LVAD infections.
Determining phage receptors and testing in animal models
In terms of biological characterization of the phages the team has already collected, next steps involve identifying their host receptors and testing phages in in vivo studies using animal models of UTIs and LVAD infections.
For now, these phage libraries are available for compassionate use in humans.
Further reading
Read the main source for this week’s feature, especially to see the figures (and much more data than I’ve described here!). Their discussion section is also particularly worth reading; in it they discuss how they envision phage therapy working in their local centers, and how their phage libraries might optimally be used.
Gibson, S. B., Green, S. I., Gu Liu, C., Salazar, K. C., Clark, J. R., … & Ramig, R. F. (2019). Constructing and Characterizing Bacteriophage Libraries for Phage Therapy of Human Infections. Frontiers in Microbiology, 10, 2537. doi: 10.3389/fmicb.2019.02537
Additional Resources
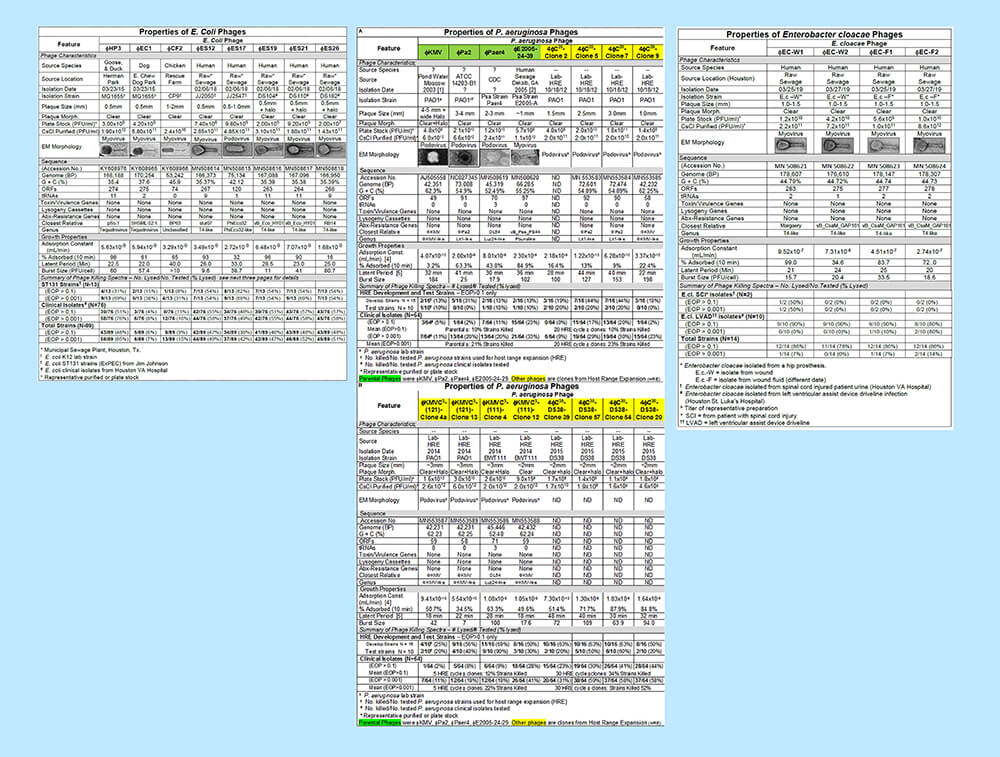
Click to view the full image of the phage characterization charts side by side (large file size). Image adapted from Gibson et al. 2019.