We’re excited to share a sneak-peek into our progress toward creating a global phage catalogue and search tool! At the start of 2020, we laid out our goals for helping the phage community manage and mine its phage data. Since then, we’ve been working hard to build a comprehensive, searchable directory of phages that can streamline access to phages and phage data.
We’re building a system that can index the phages available at existing phage collections, like DSMZ and NCTC, as well as the phages available at research labs, and the phages published in the literature. Our goal is to make it easier for you, phage researchers and phage professionals, to find the information and the phages you need.
Collaborating with the phage community
Over the past few months, we’ve started to collaborate with labs and phage collections around the world to establish a schema, which helps us understand how each collection manages the data around their phages (shoutout to BCM Tailor Labs, NCTC, and the Felix d’Herelle Center for sending us your phage metadata so far—thank you!).
Today, we’d like to share an update on how things are going, and to gather feedback from the wider phage community! Since our tool is meant to help phage researchers and professionals quickly access phages and phage data from around the world, we’d like to know what phage attributes matter to you.
Why are we doing this?
We want to make it less cumbersome and time-consuming to work with phages. We want to make it easier to find the right phage for any application or research question, and we want anyone to eventually be able to mine the database to get new phage insights.
By streamlining access to phage data, we hope to take some of the burden off of you, so you don’t have to waste hours looking up phages through reading dozens of papers and writing dozens of emails that take weeks of back and forth. An up-to-date, searchable phage tool that lets you find and reference phages quickly, and lets you order the phage from a reputable phage collection to boot, should save many hours of drudgery.
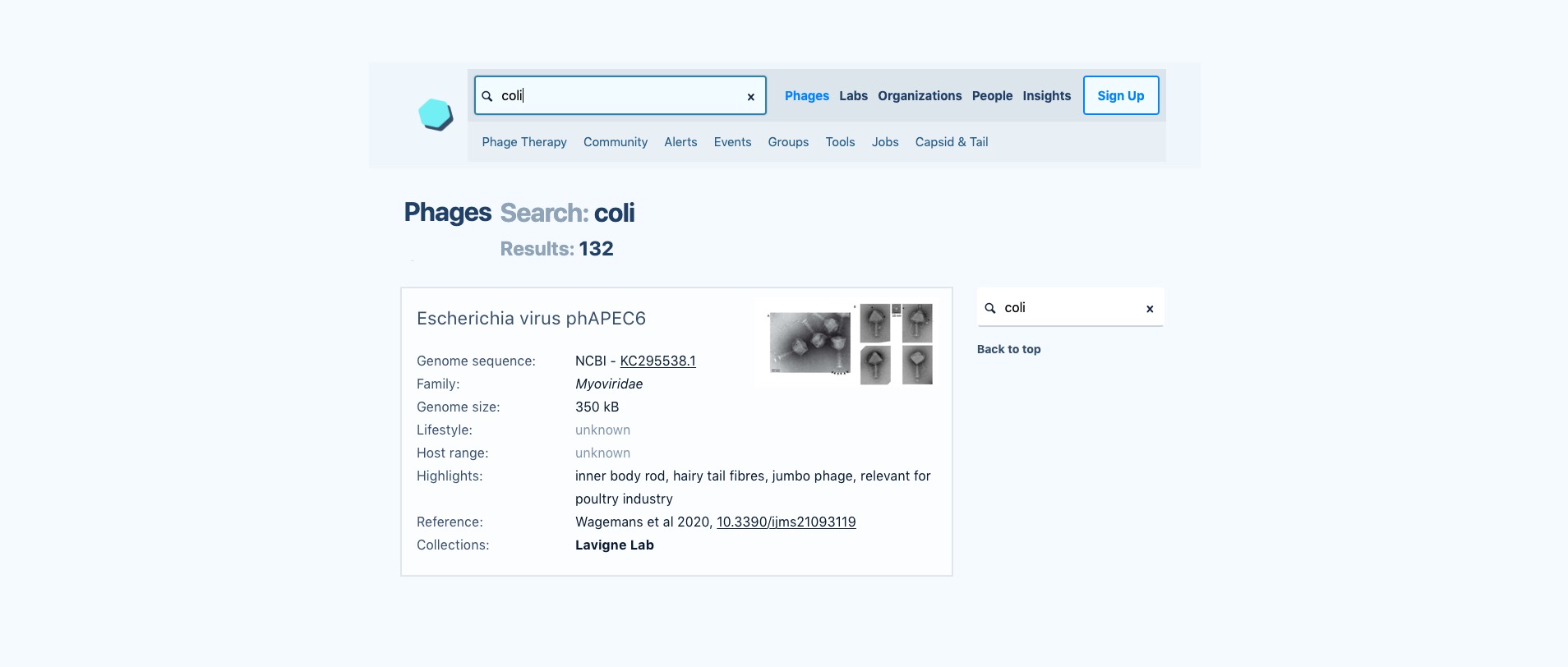
Imagining what a universal phage search tool might look like
Here’s a mock-up of what we’re envisioning a phage search might look like on the next version of Phage Directory. Many details still need to be worked out, and a lot of that will depend on the feedback we get from community members. How can we make our database and interface maximally useful for the phage community?
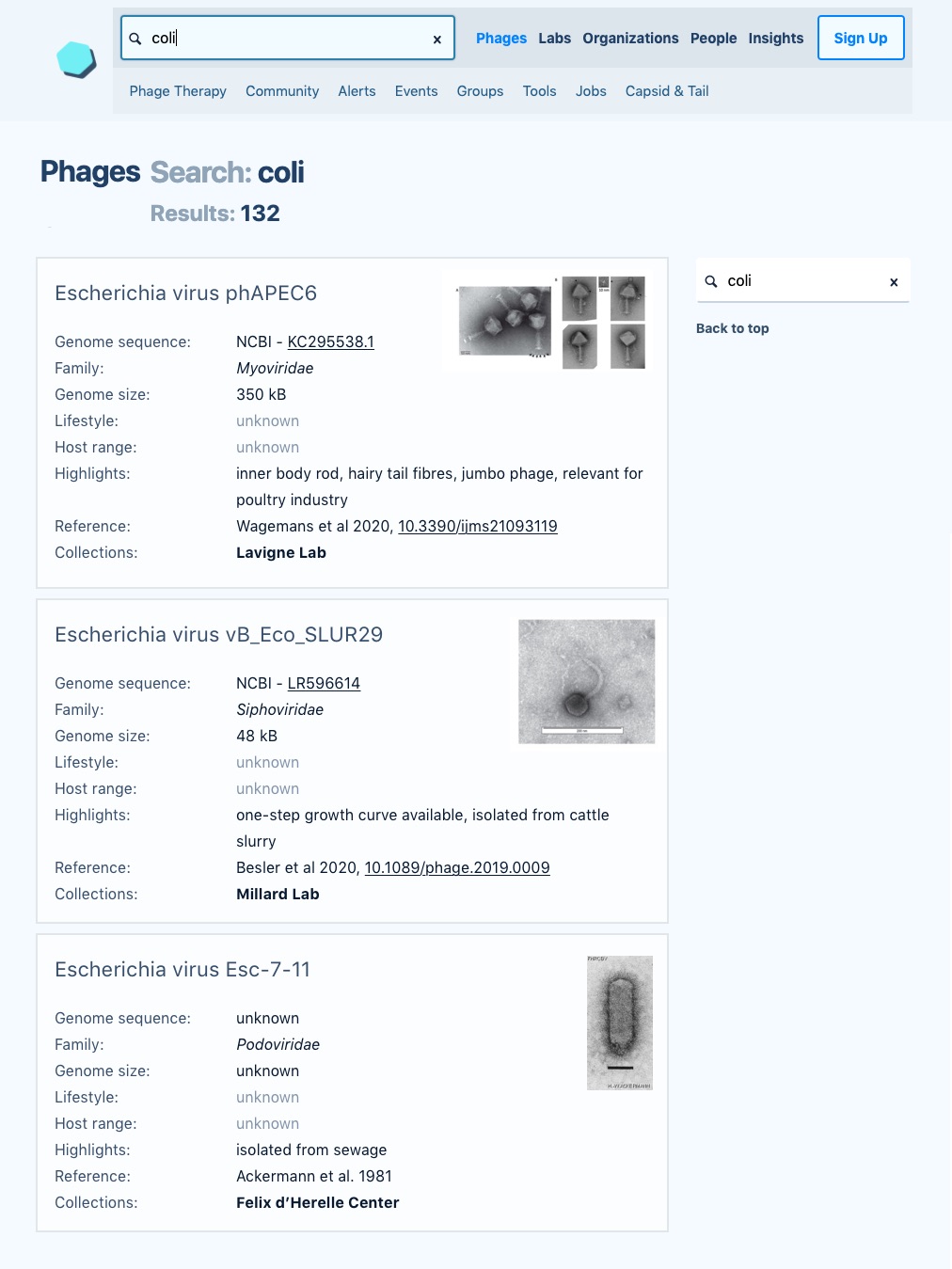
This wireframe shows an example of what we’re envisioning for the next version of Phage Directory, where we’ll be able to support searching for phages across labs and phage collections around the world using a single system. (High-resolution version.)
Building the back-end: a structured database that accepts data from any collection
To build a universal search system for phages, we need to establish a way to place each phage collection’s data side-by-side and match up one collection’s attributes to the equivalent collected by another. Because although many collections collect overlapping data, each collection/organization is driven by a different mission, and thus cares about slightly different aspects of a phage. And even when the same attribute is collected by two different collections, it’s almost certain that each will have its own name for that attribute.
For example, a piece of information like “Isolation Source” could go by many different names, like “Original source”, “Isolation”, “Source (type of sample)” or “Discovery notes”. These kinds of differences makes it really hard to make a universal phage search tool user-friendly.
Creating a master list of phage attributes
While we could build a rudimentary text-parsing system to try to tie data together, we decided to create a schema, or dictionary, that defines all the types of data that could be useful to phage research, phage therapy and other applications. Then, by mapping each collection’s list of attributes to this schema, we’ll essentially be able to translate the terms into one language. In the end, a single search term will “speak the language” of many different collections, and will bring back the relevant search results.
After combing through the metadata for several phage collections, we’ve created the beginning of our schema. We’ve divided the list into a few major categories, and given a bit of insight into what fits into each:
- Phage Identification (this one’s “easy”: what’s the phage’s name?)
- Phage Isolation (what were the circumstances of this phage’s initial isolation?)
- Phage Propagation (how should you propagate this phage?)
- Physical Characteristics (what does the phage look like? what about its plaques? how stable is it?)
- Genomic Characteristics (is the genome sequenced? what does it show?)
- Phylogenetic Characteristics (where does this phage fit taxonomically?)
- Biological Characteristics (burst size, host range, etc.)
- Reference Publications (original reference and others that build on it)
We’d love your input on what we’re collecting
We want to ensure that when we make our phage search tool publicly available, that it is actually useful for people. So we’d like to know:
- When you buy a phage or request a phage from a lab, what do you wish to know about it?
- Is there an attribute you wish labs or collections would track about a phage, but don’t?
- What aspect of the process of requesting or buying phages could be improved?
- What questions should we be asking you that we aren’t?
To share your answers, please join our Slack group, where we’ve set up a channel called #phage-metadata just for this discussion. Or, you can always email us at [email protected].
Ready to add your phage collection’s metadata?
For those of you in a position to share your lab or organization’s phage metadata with us, we really want to hear from you!
Send us an email at [email protected], and we’ll invite you to complete the phage attribute mapping exercise we’ve set up.
Rohit Kongari helped us produce this week’s article by helping us source and write the What’s New and Jobs sections. Thanks Rohit!!
Interested in becoming a Phage Directory volunteer?
Email [email protected].