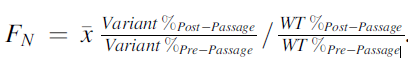This post is based on a paper by Phil Huss and colleagues (2021) entitle ‘Mapping the functional landscape of the receptor binding domain of T7 bacteriophage by deep mutational scanning’, published in eLife Mar 9, 2021.
How ORACLE works
For the development of phage therapy, what makes a bacteriophage able to infect a bacterium is an important question. The study of the receptor binding protein (RBP) on the phage helps to increase the understanding of how certain phages are effectively infecting certain strains of bacteria and not others. Huss et al. have developed an effective process of creating many different RBP variants to determine each residue’s function and importance while developing a greater understanding of the sequence-function relationship. This process, known as Optimized Recombination, Accumulation, and Library Expression (ORACLE), uses created T7 acceptor phages that have the tail fiber locus modified by adding CRE recombinase sites which flank the region.
Optimized recombination has the phages infect E. coli which have plasmids that constitutively produce CRE as well as having a tail fiber locus variant flanked by CRE recombination sites. Once the phage genome is in the cell, site-specific recombination occurs, and the tail fiber locus variant is inserted into the phage genome. This occurs many times with each E. coli plasmid with a tail fiber locus variant being slightly different with single amino acid substitutions of all 19 non-synonymous and 1 nonsense substitution at each codon spanning residue position.
These recombined phages are accumulated by enriching them through the elimination of non-recombined phage genomes. This occurs when progeny phages infect E. coli with a Cas9 and gRNA that targets the fixed sequence introduced into the original acceptor phages. The Cas9 gRNA cuts the non-recombined acceptor phage sequence ensuring only the recombined phages are able to replicate and accumulate.
These phages are then free to replicate on E. coli and create a full library of different variants where the genomes can be sequenced and the distribution of these sequences analyzed for use in studying important variants on the RBP on the tail fiber. The library of phages infected the 3 E. coli hosts BL21, BW25113, and 10G. After 4 infection cycles, variants were given scores based on the variants’ relative abundance after selection to before infection. Variant effectiveness was calculated using the formula shown:
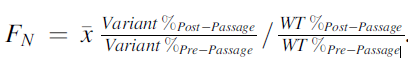
Figure 1: Formula for calculating FN for determining whether the substitution was intolerant, tolerant, or functional. Source: Dylan Woolsey
WT are wildtype T7 phages that the variants phages are based on. If FN was less than 0.1 (less than 10% as effective as wildtype) the variant was considered to be depleted. If FN was between 0.1 and 2 (10% to twice as effective as wildtype) the variant was considered to be tolerated. If FN was greater than 2 (twice as effective as wildtype) the variant was considered to be enriched. The results of the variant effectiveness can be viewed on the heat map where darker reds represent amino acid variants at a position on the tail fiber RBP that are more enriched.
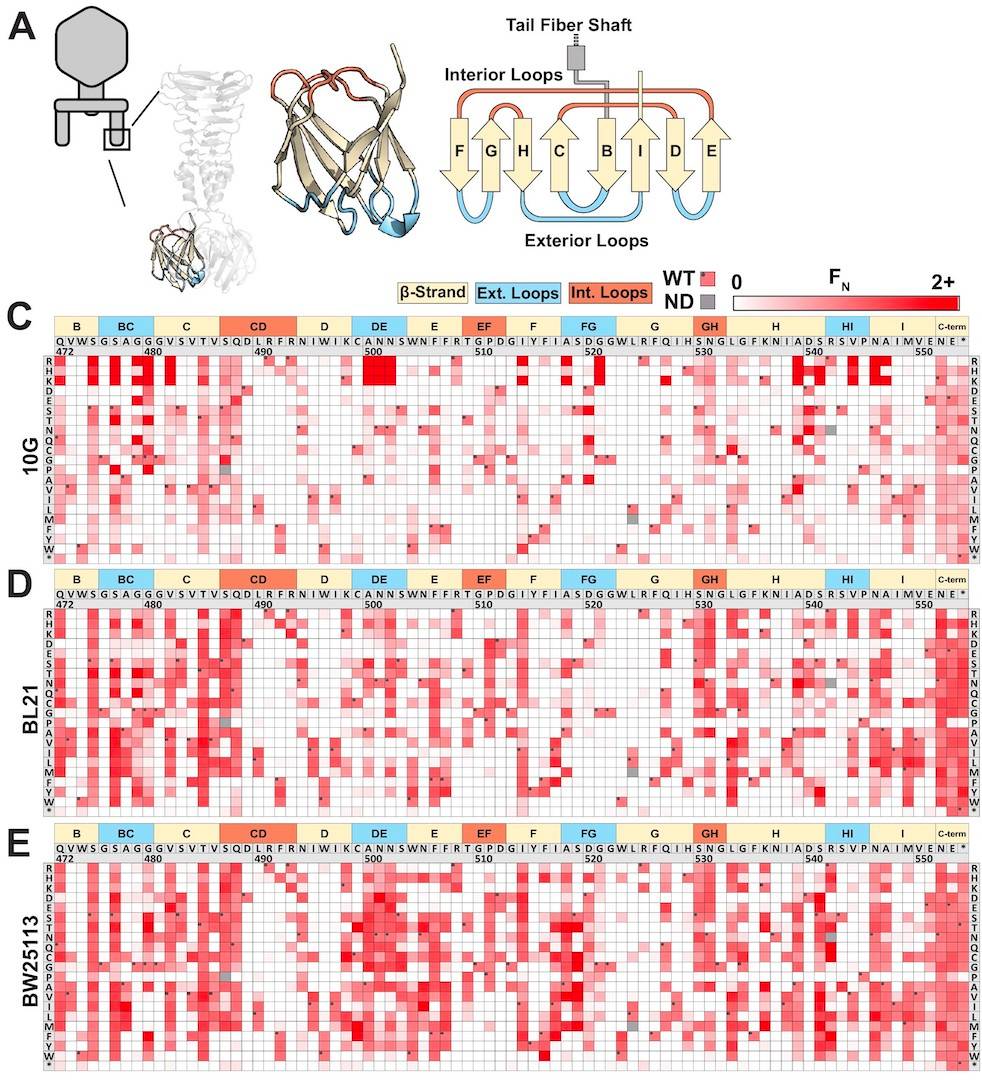
Figure 2A: The structure of the tail fiber region. 2B: Heat map showing FN values of amino acid substitutions at each position along the tail fiber. Edited from original graphic. Source: https://elifesciences.org/articles/63775
The uses of the results created by ORACLE
Important deductions can be made from these results. For instance, there is a tradeoff between effectiveness on a host and host range. This is further supported by the top enriched variants for each host being very different from one another.
Other structural information about what contributes to the ability to bind to bacteria can be gained from these results, such as there was depletion of hydrophobic amino acids and enrichment of large and hydrophilic amino acids on variants infecting 10G.
Besides broader information about variants, more specific information can be found from the results. For example, exterior loops in the tail tip have a greater proportion of functional substitutions (ones that are depleted in 1 variant and not in another) seen in the figure below where also shown are intolerant variants (being depleted in all hosts) and tolerant variants (being tolerated in all hosts).
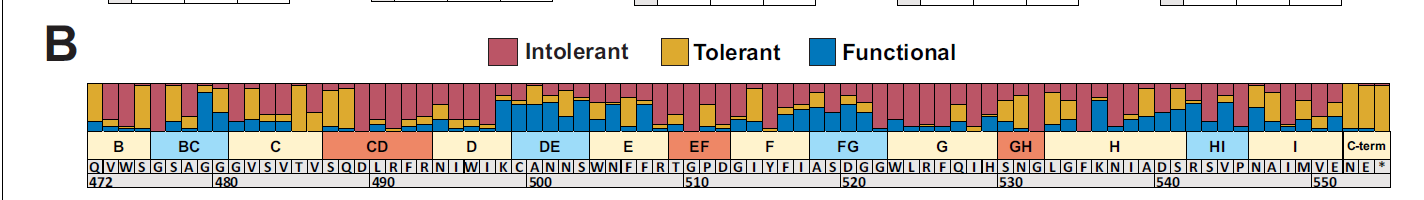
Figure 3: Proportion of each position on the tail fiber having intolerant, tolerant, and functional substitutions. Source: https://elifesciences.org/articles/63775
In addition to the addition of structural knowledge gained through ORACLE, the use of the library and subsequent ability to characterize each variant has been proven to be valuable tools in combatting resistant bacteria. When the researchers deleted LPS regions of bacteria, they were able to use the library to find variants able to infect these strains. As well, with a UTI E. coli strain known to develop phage insensitivity rapidly, using the library the team observed insensitivity was greatly delayed and bacterial load was decreased compared to wildtype.
From these tests, specific effective variants were also found. Since these are known variants, more information about effective tail tip RBPs can be learned by studying what is unique structurally about these variants.
Overall, the use of the ORACLE method provides the ability to learn more about phage variants structurally through amino acid changes as well as helping develop a better understanding of what makes effective bacteriophages for phage therapy.
Further Reading
Phil Huss, Anthony Meger, Megan Leander, Kyle Nishikawa, Srivatsan Raman (2021), Mapping the functional landscape of the receptor binding domain of T7 bacteriophage by deep mutational scanning, eLife 10:e63775.