Hi everyone,
This week I got to tackle a passion project that I’ve wanted to do for a really long time, which I call the “Open Phage Data Sheet.”
Despite the fancy name, the premise is simple: it’s a publicly editable spreadsheet of phage resources like grants, clinical trials, and bioinformatics/lab tools!
There are several tabs/sections in the spreadsheet, as listed below:
Section |
Description |
| Bioinformatics |
Both viral and bacterial bioinformatics tools, and various resources like benchmarks and tools lists. |
| Clinical Trials |
A running list of clinical trials, mostly pointing to https://clinicaltrials.gov. Would be nice to have old, historical trials, plus case studies and similar. |
| Events |
Upcoming webinars, conferences, meetups, with dates, location, etc. Both Paid and free. |
| Lab Tools |
Tools for the wet lab, from pipettes to liquid handling robots. And references to benchmarks of “cheap Amazon lab stuff” that might actually work! |
| Definitions |
Glossary of terms. These can be “subjective” to the author, and multiple definitions are more than welcome (based on context, usage, language, etc.) |
| Grants |
Public or private grants for phage, bioinformatics, and adjacent fields |
| Resource Library |
Links to other resource lists, databases, search engines, comparison papers, phage journals, and generally useful links. Bonus points for actively managed resource lists |
| Funding Opportunities |
Awards, travel grants, studentships, and that sort of thing |
| Contributors |
Volunteers who contribute to the Open Phage document |
For now, many of these tables are empty (I just started this project) but I hope to fill in these details over the coming weeks and months — hopefully with the help of the phage community!
Here’s the link to the full Google Sheet: https://open.phage.directory/sheet
A bit of background
This is heavily inspired by many existing open sheets projects, like Nouri’s (our Phage Australia bioinformatician) Phage Kitchen, and fellow Aussie Rob Edwards’ Viral Bioinformatics Tools, or Pieter Levels’ Nomadlist, which started as a Google Sheet.
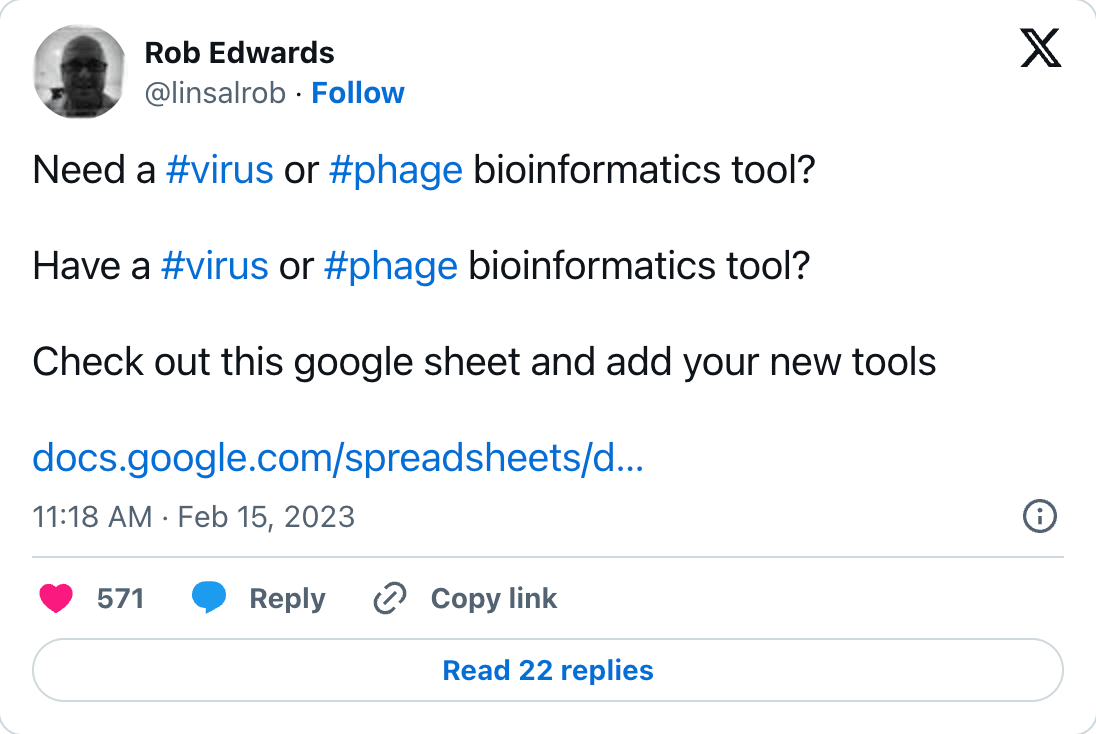
Fig 1. Rob Edwards’ tweet about his open Google Sheets bioinformatics tool: https://twitter.com/linsalrob/status/1625650675236454400?s=20
For the many years we’ve been doing Phage Directory, we’ve always used Notion and Airtable for Community Board work. I did this mainly because they play more nicely with web code — which is how Phage Directory and Capsid & Tail gets all their data.
The problem with Notion and Airtable is that they’re a complete pain for the community to edit — both require sign-ups and sign-ins to edit “publicly-editable” documents. Most people don’t really have Notion or Airtable accounts, which makes this a huge hassle for many people. This eventually leads to me as the sole curator of many projects — which is simple but difficult — as the curator needs to be consistently involved.
We tried to do something like this for bioinformatics a while ago, during the pandemic: https://bioinfo.phage.directory. It meant well, but the difficulty of adding information meant it wasn’t really going to go anywhere. It became a burden for me to constantly look for new papers and add new tools to it; especially since I’m so new to bioinformatics myself, I didn’t really know how to categorize and manage new tools.
For our other endeavors like finding news, jobs, and community posts, our amazing volunteers like Atif have been holding down the fort and doing amazing work finding and cataloguing them over the years. I thought it would be neat to have a similar place for longer-lasting information like tools, grants, and other resources — all co-created by the community!
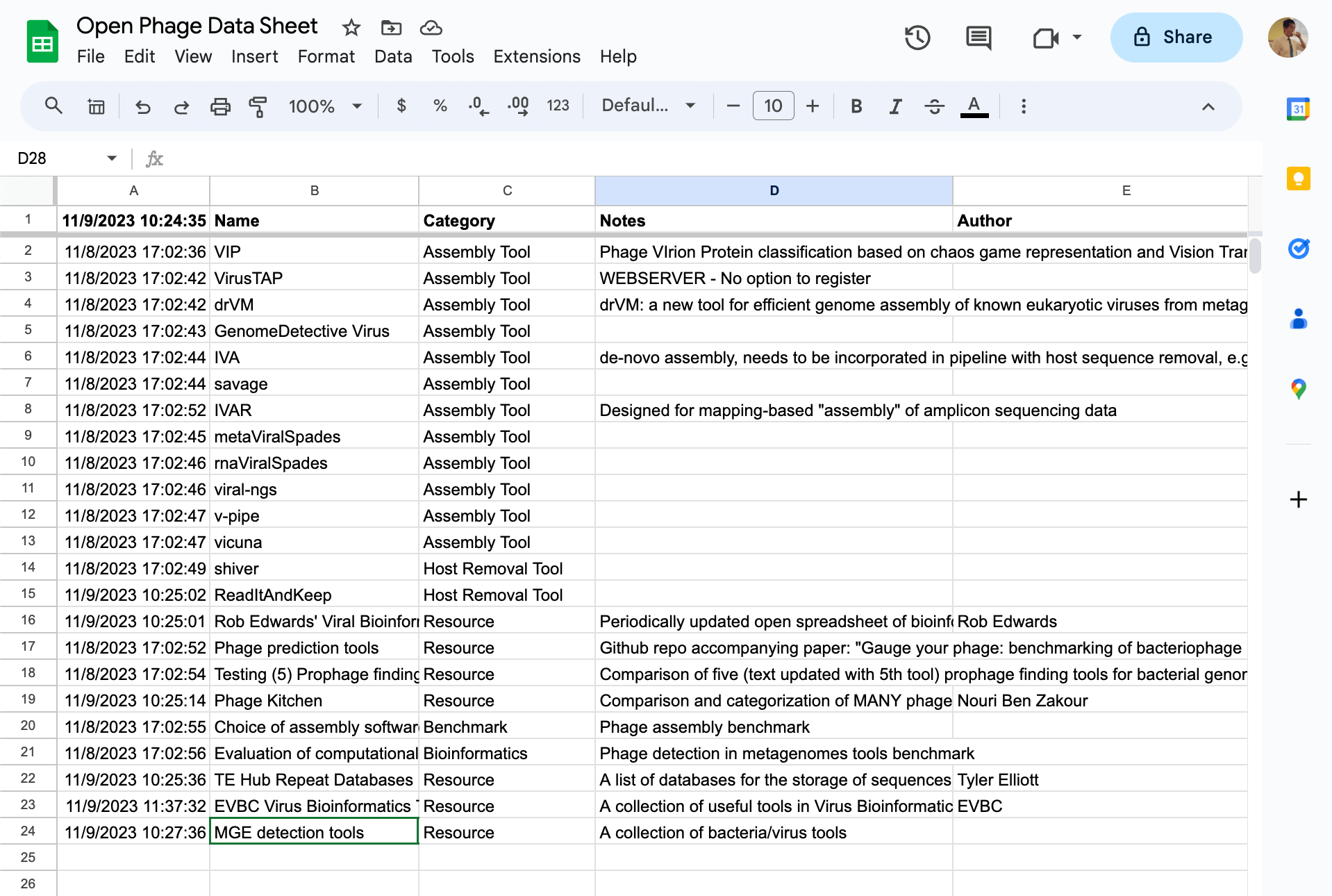
Fig 2. The Bioinformatics Tab in the Open Phage Data Sheet (https://open.phage.directory/sheet)
Going beyond bioinformatics
I love Rob Edwards’ viral bioinformatics tools Google Sheet — his Sheet is also open and co-created with the phage community — but unfortunately it’s only for bioinformatics! We’re adding a few more sections in hope that we’ll be a resource for more than just bioinformatics.
In addition to the Google Sheet, we’ve built a project page, which can be found at: https://open.phage.directory, which pulls together the data from all the tabs into a single page.
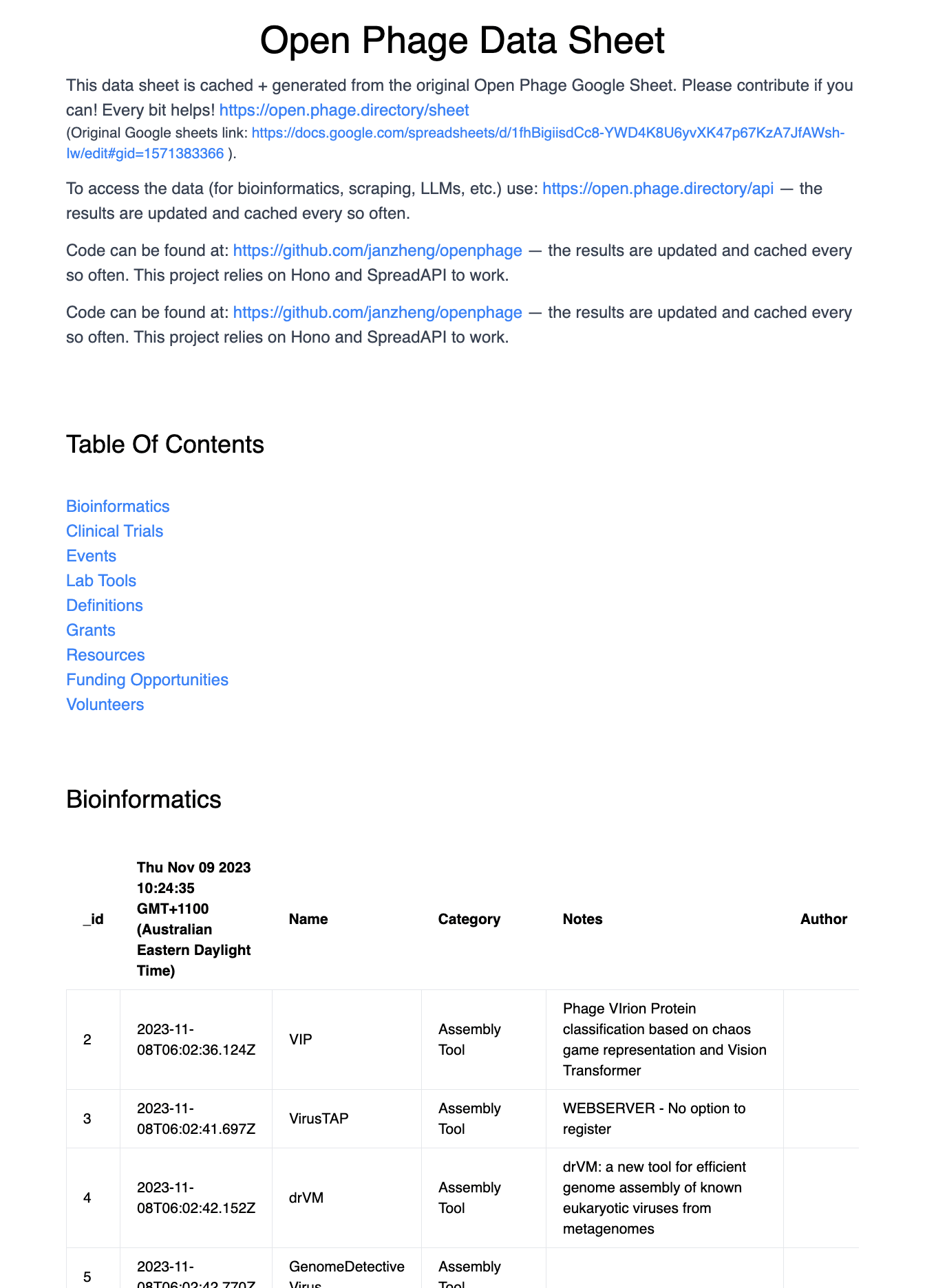
Fig 3. I built a simple project page to aggregate the data from all the sheets (https://open.phage.directory)
This Project Page is hopefully a useful aggregated view of each table in the spreadsheet.
Ok so I admit, the project has lot of empty sections (I did start this project a few days ago) and I’ll try add more content, though any help is appreciated.
I’m especially keen on adding to the “Resource Library” tab as it will feature other resource lists, databases, search engines, and so on — essentially places where I’ll find more items to add to this spreadsheet.
Over time, I can think of more sections to add over time, like “Phage Projects” like Vivek’s Phage Foundry or Tobi and Fran’s "Developing Scientific Guidelines for Phage Banks” effort, or Networks like Phage Australia, Phage Canada, or Phage UK / UK Phage.
If you’d like more sections, feel free to add them and email me ([email protected])!
Project Goals
Phage Directory was always created with the mission of being a “one stop shop for phage researchers.” It’s been growing over time, is buckling under the weight of heavier-than-expected use and signups (the current code is almost 5 years old!). This means I’m exploring alternatives to the current architecture — like Google Sheets!
It might sound like this project is in direct competition to the Phage Directory (uses Airtable forms for data entry). But I wanted to create something fully open (everything is editable by anyone) to really lower the barrier of resource sharing. This is an experiment in complete openness of co-creation!
Success for this project looks like the community making this project their own, and regularly adds their own resources and tools.
Beyond the sheet
Fig 4. Open Phage is open source — feel free to create your own system (https://github.com/janzheng/openphage)
Over time, I’m exploring ways to automate adding these resources alongside the community, for example with AI tools that can extract information from research papers.
I’d love for Open Phage to become a single place to easily search for “what you need” across the phage field, like what tools to use, what grants to apply for, etc.
For now, the project page provides a simple API for all aggregated data: https://open.phage.directory/api.
Eventually, a search tool for Open Phage could live on Phage Directory, alongside the current areas like Profiles or Capsid & Tail.
Under the hood
For bioinformaticians and techies, I built a tiny microsite using Cloudflare Workers and Hono, a JS server framework that pulls data from the Google Sheet. This mainly serves as an aggregator and a simple demo of what can be done (and as a way to copy/paste the data from all tabs into a GPT, Assistant or Claude). The Google Sheet itself uses a free App Script called SpreadAPI that turns the Sheet into a fully-featured REST API endpoint! What that means is that you can get data from any tab (as well as update or create new rows…!) using another computer, server, or app.
This is especially powerful in the age of AI where any information can be retrieved from any of these resources. Very powerful tools can be built on top of these links!
The API itself that pulls data from ALL the tabs is available as a JSON GET endpoint here: https://open.phage.directory/api. The data from the Sheet is cached using Cloudflare Workers KV and is set to expire every 24 hours (because of Google’s aggressive rate limits).
If you’re a builder — once more content is added — you can build all kinds of tools on top of this resource: a grants aggregator, summarizer, and auto-grant-writer; a data analysis and code interpreter tool that selects, downloads, and runs each bioinformatics tool; a Q&A tool on top of any research paper or grant. The things you could build are endless!
Volunteers Wanted!
Over the next few months, I’ll be slowly adding more information to the sheets as I come across them, while monitoring for malfeasance (it doesn’t seem like Rob Edwards’ list has received very much of that).
But any help is appreciated! If you’d like to volunteer, just hop on over to the sheet and start adding: https://phage.directory/sheet.
Don’t forget to add your name to the “Added By” column (so you get the credit), and add yourself to the Volunteer tab.
Remember that this is a public, open community resource meant to help everyone, so please keep it civil!
Email me if you have any ideas or want to add more sections: [email protected]
Happy Open Phaging!
~ Jan