For this week’s feature article, Dr. Rohit Kongari spoke with Dr. Vivek Mutalik, a scientist from Lawrence Berkeley National Laboratory (LBNL), Berkeley, CA, USA about his journey to become a phage researcher and how he is using his engineering background and penchant to develop tools to answer questions in phage biology.
The discussion is largely based on the following two preprints published by the Mutalik lab, Deutschbauer Lab and Arkin Lab at LBNL:
- High-throughput mapping of the phage resistance landscape in E. coli
- Systematic Discovery of Salmonella Phage-Host Interactions via High-Throughput Genome-Wide Screens
Rohit: Could you give us a little background about your career path to being a Research Scientist at the Lawrence Berkeley National Lab?
Vivek: I am originally from Belgaum, a small city near Bangalore, India. I did my undergrad in pharmacy in Belgaum and then I went to University Institute of Chemical Technology (known as UICT) at Mumbai, India to pursue a master’s degree in Bioprocess Technology. Having developed an interest in large-scale fermentation, protein production and purification, I wanted to explore the fermentation industry.
Over the next 3 years, I worked in supervisory roles at a couple of fermentation companies, overlooking the large scale manufacturing of antibiotics and vitamins, like Penicillin and B-12. Quickly realized that research is something I really enjoy, and I returned to academia to do a Ph.D. in Biochemical Engineering from Indian Institute of Technology (IIT) Bombay in Prof KV Venkatesh’s lab.
My PhD work was focused on computational analysis of enzyme cascades, switch-like responses in signaling systems and ways to uncover emergent properties of genetic regulatory circuits. It was when I joined Prof. Carol Gross’s lab at UCSF as a postdoc that I really started getting hands-on experience in a microbiology lab with the help of my mentor and friend Dr. Virgil Rhodius.
After four years of valuable learning experience in bacterial stress response and transcriptional regulation at UCSF, I then joined Lawrence Berkeley National Lab as a project scientist to work with Prof. Adam Arkin. It was a position that allowed me to work on some of the exciting ongoing synthetic biology projects (in collaboration with Prof Drew Endy from Stanford Univ. and Prof. Jay Keasling from UC Berkeley and LBNL) such as BIOFAB funded by National Science Foundation. I have now moved up to the position of a Principal investigator, a full-time career research position with my own group within the Environmental Genomics and Systems Biology division and the Biological Systems and Engineering Division of LBNL.
R: When and how did your interest in phages start?
V: Throughout my research career, I have always been passionate about developing tools and technologies to study and engineer bacteria for applications, and to uncover regulatory mechanisms and gene functions. Even now (after decades of research work in genetic engineering/synthetic biology), researchers do not have access to tools to genetically modify most non-model bacteria. My team and I are passionate about developing such tools for non-model organisms important in diverse biotech applications. While we were developing a couple of technologies to examine gene expression and function with an intention to study bacterial genes with unknown function, we quickly realized that these tools were apt for phages too.
For an outsider to the field of phage biology, it is easy to get overwhelmed with the abundance of interesting questions to ask and it can be really challenging to know where to start. However, we were fortunate that the tools that we developed fit into the existing technological gap to study host-phage interactions in a high-throughput manner in diverse environmental contexts.
The more I learned about phages, the more I got fascinated with their biology and found a lot of interesting questions that can be addressed with genetic tools we have developed. We eventually chose to focus on understanding host resistance to phages and associated evolutionary trade-offs, in addition to developing tools for phage functional genomics and engineering, given the current relevance to antimicrobial resistance and reemergence of phage therapy.
R: Could you briefly describe these tools and/or approaches that are currently being used in your lab and their range of applications?
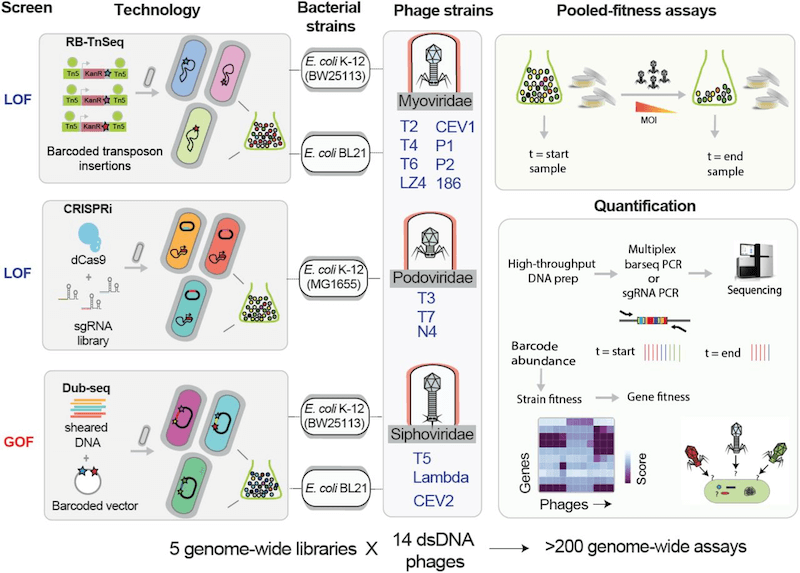
V: Our recently published work has been a product of three different approaches (RB-TnSeq, CRISPRi and Dub-seq) used in answering the question of host gene function. These three high-throughput genetic technologies enable fast, economical and quantitative genome-wide screens for gene function, which are suitable for discovering host genes critical for phage infection and bacterial resistance.
First, the Random barcode transposon site sequencing (called RB-TnSeq) allows genome-wide insertion mutagenesis leading to loss-of-function mutation; in this approach, genome-wide transposon insertion mutant libraries labeled with unique DNA barcodes are generated, and next-generation sequencing methods are used to map the transposon insertions and DNA barcode at loci in genomes; second, CRISPR interference (CRISPRi) approach, which allows partial inhibition of gene function via transcriptional inhibition. In this CRISPRi approach, a catalytic null mutant of the Cas9 protein guided by chimeric single-guide RNA is used to programmably knockdown gene expression, thereby allowing the probing of all genes (including essential genes) and more precise targeting of intergenic regions. Both these tools have their own pros and cons; Although RB-TnSeq can be applied on a large scale across multiple bacteria through barcode sequencing (as shown by my collaborators, Adam Deutschbauer and Adam Arkin labs, LBNL), it is limited to non-essential genes, whereas CRISPRi can be used to examine essential genes, but the design and Cas protein expression optimization stages require a lot more effort and time.
In addition to these two loss-of-function approaches, we have developed a gain-of-function method called Dub-seq or Dual-Barcoded Shotgun Expression library Sequencing, which queries the effects of gene dosage or gene over-expression in parallelized fashion. Essentially, Dub-Seq uses shotgun cloning of randomly sheared DNA fragments of a host genome on a dual-barcoded replicable plasmid and next-generation sequencing is performed to map the barcodes to the cloned genomic regions.
Intriguingly, the effect of gene dosage or gene overexpression on phage growth and phage resistance has not been well studied for most phages even for model E. coli strains. It has been observed quite often that antibiotic-resistant clinical isolates have gene duplications or mutations amplifying gene expression, so it made sense to us to check if we could see similar patterns in phage resistance phenotypes. While our initial results from Escherichia and Salmonella have been published as preprints recently, we are currently in the process of building libraries in other important pathogens such as Pseudomonas, Acinetobacter and Klebsiella.
Many interesting questions about host physiology and host-phage interactions in these model systems have not been addressed so far due to lack of proper genetic tools and we hope to bridge at least some part of it through our large-scale approaches. As a bench researcher, one of the coolest things about these genetic screens is the ease of experimentation in diverse contexts at much cheaper cost. Of course, building these genome-wide libraries need an upfront investment. For example, building a typical genome-wide library costs up to $4000 to $8000 and 3-6 months of time, but once the library is built, it serves as standardized genetic tool kit that can be used innumerable times for addressing interesting questions.
R: Talking about your preprints, could you highlight a few of the interesting observations from your work on the phage resistance landscape in E. coli and Salmonella?
V: When we started this project, our initial idea was to generate a proof-of-principle dataset for our new technologies using classical phages. Thus, we started with a couple of basic lab E. coli strains (MG1655 and BW25113) and a set of 11 canonical phages (like T1 to T7, P1, N4) alongside three relatively new phages. Given the ample amount of literature on host factors involved in the growth of the well-studied phages, we were not really expecting to uncover new targets. However, these high throughput approaches brought to light some interactions, that may have been missed or overlooked in classical genetic approaches. For example, even for a phage as well studied as T4, we were able to find an E. coli gene with no previous annotated function (ygbE) that conferred phage resistance upon overexpression. Upon further analysis, we understood that phage resistance was an indirect effect of the downregulation of OmpC, the surface receptor for T4.
One more unexpected find from that E. coli study was the role of cyclic-di-GMP in infection by phage N4. Our data showed that overexpression of multiple different host factors involved in the cyclic-di-GMP pathway conferred resistance to phage N4. However, the transcriptional levels of the N4 surface receptors (NfrA and NfrB) were unperturbed in this case, so there seems to be a different mechanism affecting the infection pathway which needs more follow up studies.
Another noteworthy observation was the widespread effect of colanic acid biosynthesis on phage sensitivity. Very early studies in E. coli and some of the most recent reports in Pseudomonas had proposed that the mucoid phenotype of mutants with elevated colanic acid expression probably interferes with receptor accessibility for phages. Our data shows that this could be a more generalized form of host defense that could be extended against a diverse set of phages.
The Salmonella study was done using a lab strain MS1868 and a set of 11 phages, but only 4 of the 11 phages we tested had previous substantial studies done on them. The results from those experiments not only threw light on novel adsorption strategies but also gave in-depth information about the LPS structural requirements for adsorption of diverse Salmonella phages. The other highlight of that work is finding the involvement of the sigma factors RpoN and RpoS in mediating phage resistance, with the latter being able to confer cross-resistance to multiple phages. Specifically, the role of virulence-associated RpoS on phage resistance is intriguing and needs detailed follow up.
R: I noticed that all your current work focuses on pathogenic Gram-negative bacteria, do you plan to employ these tools to study phages of Gram-positive bacteria?
V: Yes, we are working on Gram-positive hosts too, but there are some technical difficulties we are trying to address. For example, one of the challenges for our approach is getting a significant number of colonies from a transformation reaction sufficient enough to generate a diverse set of bar-coded strains. The current limitation in transformation efficiency or the general genetic tractability of Gram-positives is the major hurdle for us in library preparations and we are making efforts to develop generalized genetic tools to overcome that.
R: What kind of implications do you think these tools and high throughput approaches hold for phage therapy and other applications?
V: I think these tools will have a huge impact on phage therapy, especially the aspect of formulation of phage cocktails for therapeutic purposes. All the collateral sensitivity and cross-resistance data generated using these high-throughput technologies would contribute to a rational phage cocktail design and combinatorial treatments with antibiotics. Most importantly, these technologies provide us large-scale phage-host interaction landscape dataset that can be used for developing predictive computational approaches on phage infectivity based on phage and host genome sequence alone. Also, these technologies have also been very useful to understanding the lysis mechanism used by phage lysis proteins and other phage encoded host toxic genes.
In addition to the phage therapy side, our group is trying to establish a platform where we use the knowledge gained from these new discovery-oriented tools and apply them in engineering-oriented tools. The applications could range from engineering a single phage for genetic manipulation of a bacterial host or host range expansion to formulating a phage cocktail for efficient elimination of a pathogen in a community of microbes.
Also, let us not forget that these genetic tools provide a great platform to study host biology as well. We examined host gene function using these genome-wide tools under different stress conditions such as nutrition starvation, DNA damage, and antibiotics. Our work comparing the host fitness under different stress conditions and describing host factors contributing to antibiotic resistance has been published. Now equipped with all these data available in terms of host fitness, antibiotic resistance, and phage resistance, we hope to devise better antimicrobial strategies.
To extend these tools and technologies to practical clinical applications and helping patients to treat antibiotic resistant infections, recently, I co-founded Felix Biotechnology, a company focused on accelerating the deployment of novel biotherapeutics targeting urgent microbial challenges in human health. At LBNL, we are working towards establishing the Phage Foundry, a centralized facility to connect technologies to characterize phage-host interactions and develop tools for seamless genome engineering of phages to provide knowledge and viral reagents to the broad research community focused on the agricultural, environmental and health challenges of our times. By developing the foundational knowledge on phage predation and seamless genome-engineering of phages, we’ll be able to make strides in precise microbiome manipulations and develop strategies to tackle antibiotic resistance infections.
R: Well, that was a great chat! We all look forward to more fascinating work from you and your group. Would you like to add any concluding remarks?
V: Thanks for the great chat. I would like to say that I have definitely benefited by being at a place like Lawrence Berkeley National Lab, surrounded by a great interdisciplinary team of graduate students and postdocs, and awesome collaborators and friends. Partnering and long-standing collaborations with Drs. Pavel Novichkov, Alexey Kazakov, Morgan Price, Adam Deutschbauer, Adam Arkin, Britt Koskella, Kim Seed, Richard Calendar, Elizabeth Kutter, Simon Roux, Matthew Sullivan and Paul Turner has been critical, given the expertise and knowledge they bring with them. In terms of funding, my team is supported by funding from Innovative Genomics Institute, and from the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research.