Once we’ve thought about what we need to know about our phages, we can start giving them identities. How we identify our phages tells us how we should name them. Do we affectionately name them after our pets (“Rue”), or do we give them a serial number (“NCTC 12673”)? Or do we use both?
In this second post in the informal and conversational “Phage Data series”, I’ll step through how I think about naming our phages for Phage Australia.
When we think about the identity of a person, place, or thing, the first thing we think of is its name. Names are powerful — merely having a name for some thing helps us talk and think about the thing. When everyone agrees on what to call the thing, the thing gains identity and a place in the world.
For our phages, names help us think and talk about them — both from within and beyond our lab. Names help us compare them, tweet them, publish on them, and accession them into Genbank. Names help us accession our phages into collective culture.
Names are powerful, and memorable names are doubly so. How will others cite and mention our phages? How will the world talk about them when they’re used to prevent the amputation of a 7-year-old girl’s leg?
Access(ion)ing our phages
We need a name when we need to find our phage from our database, from our freezers, from around the world. When creating a phage accessioning system, I think of using a library or museum as an analogy for Phage Australia. We’re a library actively curating multiple collections of phages. How do we think about accessioning (recording new phages) into our own collection?
Phages can come from the wild (isolated in the lab) or from another lab or biobank. Phages can be evolved or engineered. They can be shared with other labs, who then evolve or engineer their own versions. Multiple, slightly different versions will exist around the world. How do we think about these phages, and how would we know how they’re related? How do we refer to our own copy of a phage, versus some other biobank’s exact copy of that phage? Are they related from a genomics or from a provenance standpoint?
How could we name our phages in a way that embeds some of this hidden knowledge?
If we (in the near future) create some inter-library loan system for phages, how should we be able to search for, request, deposit, share, and access these phages?
What’s in a name?
“That which we call a T4 by any other name would plaque just the same” — Shakespeare (if he was a phage biologist)
The primary goal of a name is communication. Whatever you choose to call your phage, make sure that everyone agrees on its name. To aid in this goal, make it memorable. “Rue” is much easier to remember than “NCTC 12673”. SEA-PHAGES lets students name the phages they find, adding a degree of inclusivity and ownership.
A phage by the name of “Kangaroo” could go viral on Twitter (or get cited in articles) much more easily, merely by being memorable. To increase citations and mentions, try to give phages names that help them track back to your lab.
At Phage Australia, we call these “common names”. Common names are easy to pronounce, easier to remember, and harder to misspell or get wrong (was it 12673 or 12637?). Easy to remember phages are also easily found. If we need to find any data for it, we just search for “phage Kangaroo”.
Starting with common names
Naming our phages can be as simple or as complicated as we want. Naming a few dozen phages should be straightforward. ICTV suggests combining isolation strain’s genus, the word “phage”, and a common name. Examples include “Escherichia phage T4” or “Mycobacterium phage Muddy”.

Using a common name like “Muddy” makes the phage memorable. It also acts as an “accession ID”. For example, here’s Muddy’s data on PhagesDB: https://phagesdb.org/phages/Muddy/
Simple common names are memorable, and great for daily use. However, if we plan on isolating/organizing hundreds or thousands of phages, giving each phage a distinct, memorable name will quickly become a chore.
When we start dealing with hundreds or thousands of phages, we will want to have a naming system. A system helps us add context to our common phages. For example, there are many people named “Jessica”. “Jessica Sacher” and “Jessica Simpson” are separated by a surname. Think of “given name” and “surname” as a naming system.
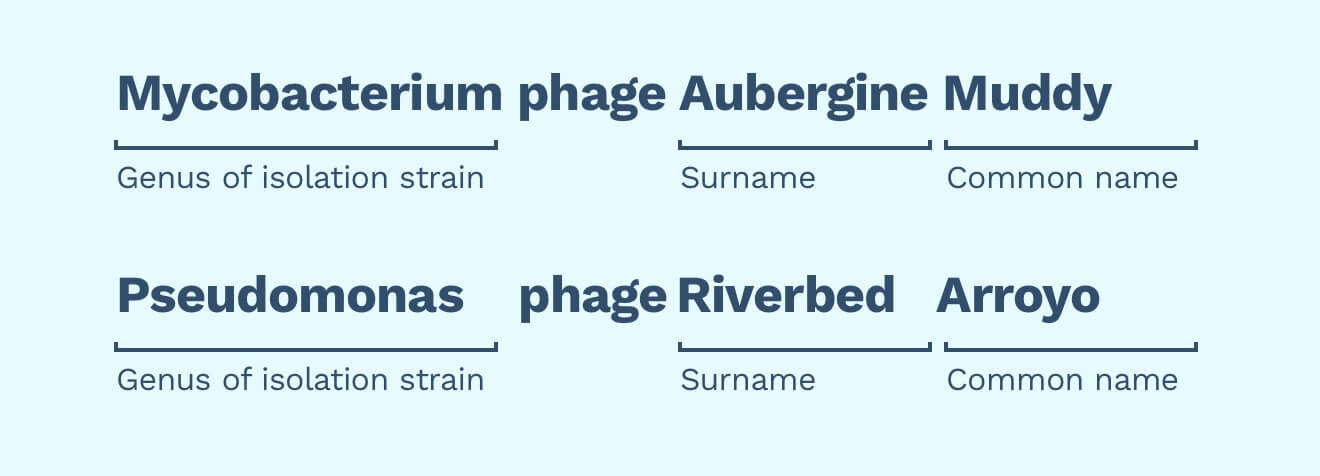
Surnames are additional names we give to people to help us set them apart. To set our own phages apart, we could also use “surnames”. Surnames can help us identify the context or heredity of our phage. For example, was the phage isolated from an aubergine, or from the bottom of a riverbed?
Our lab will soon collect and manage lots of phages, and not every phage will be noteworthy. Some of our isolated phages we might save for future characterization. For these phages, we want a system that lets us easily recall and identify them, without needing common names. We want a flexible system that lets us use isolation strain and series names, on top of an optional common name, to add more context.
Adding complexity to phage names
For larger phage collections, a more contextual naming system will help us organize the phages in both our spreadsheets and freezer boxes.
We’re thinking of a more complex “surname” system that adds more context to the name.
For example, phages named “Pa1”, “Pa2”, …”Pa1000” indicates a set or series of phages isolated on P. aeruginosa. Alternatively, “WasteWater1”, “WasteWater2”… indicates source, ie. phages isolated on waste water.
Combining the two concepts, we get “WasteWater Pa1”, “WasteWater Pa100”, “WasteWater Ec1”, … “WasteWater Ec100” — this identifies the waste water series of phages isolated on both E. coli and P. aeruginosa.
We can still use common names! We could give “WasteWater Pa5” a memorable and Australian name like “Kangaroo” — this is the name we call it in the lab, in publications, and on social media. The full database name (”accession ID”) is “WasteWater Pa5 Kangaroo”.
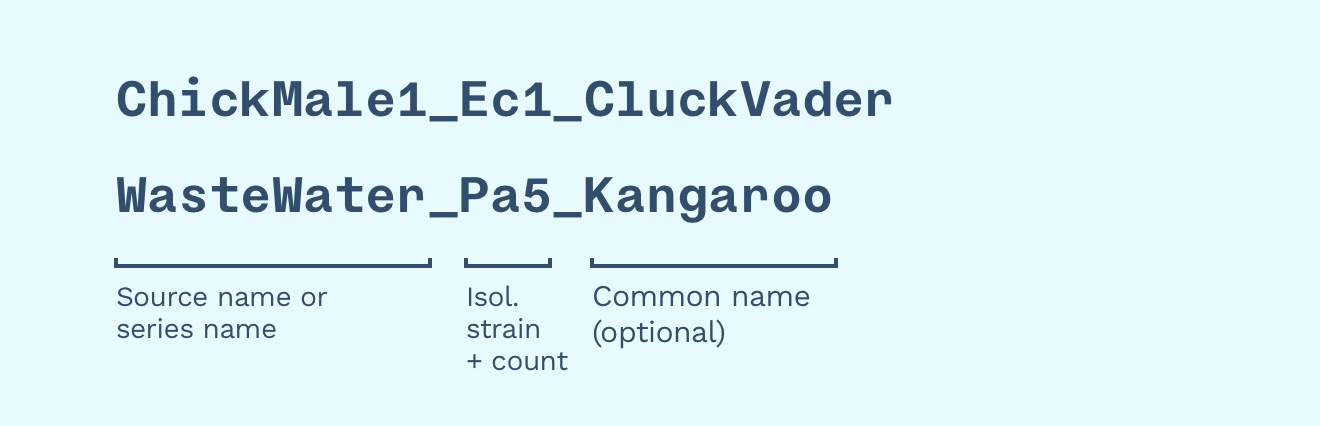
In this example, “ChickMale1” or “Wastewater” is the source or series name, “Ec1” or “Pa5” is the surname (using isolation strain and a number, e.g. “fifth phage found), and “Kangaroo” is the common name.
Not all phages would need a common name — some could just be called “WasteWater Pa5”.
If we choose to publish and accession a common name for a phage, we should always make sure to make it memorable and unique. We should also Google the name and make sure it doesn’t already exist in either phagesdb.org or Genbank.
SEA-PHAGES has a handy guide for naming phages here: https://phagesdb.org/namerules/
Choosing a more pedantic naming system
If we need other labs to interact with our bank (e.g. request, borrow, donate/deposit, stockpile/cache, or even invest in phages), we would need to choose a more flexible accession system.
We would need to accommodate other biobanks’ naming strategies. Phages also need to account for provenance by using other labs’ identifiers as “surnames”. We might also take in non-phage material, so we should indicate that a sample is a phage (”vB”, as “virus of bacteria”, as opposed to a lysin, synthetic phages, etc.).
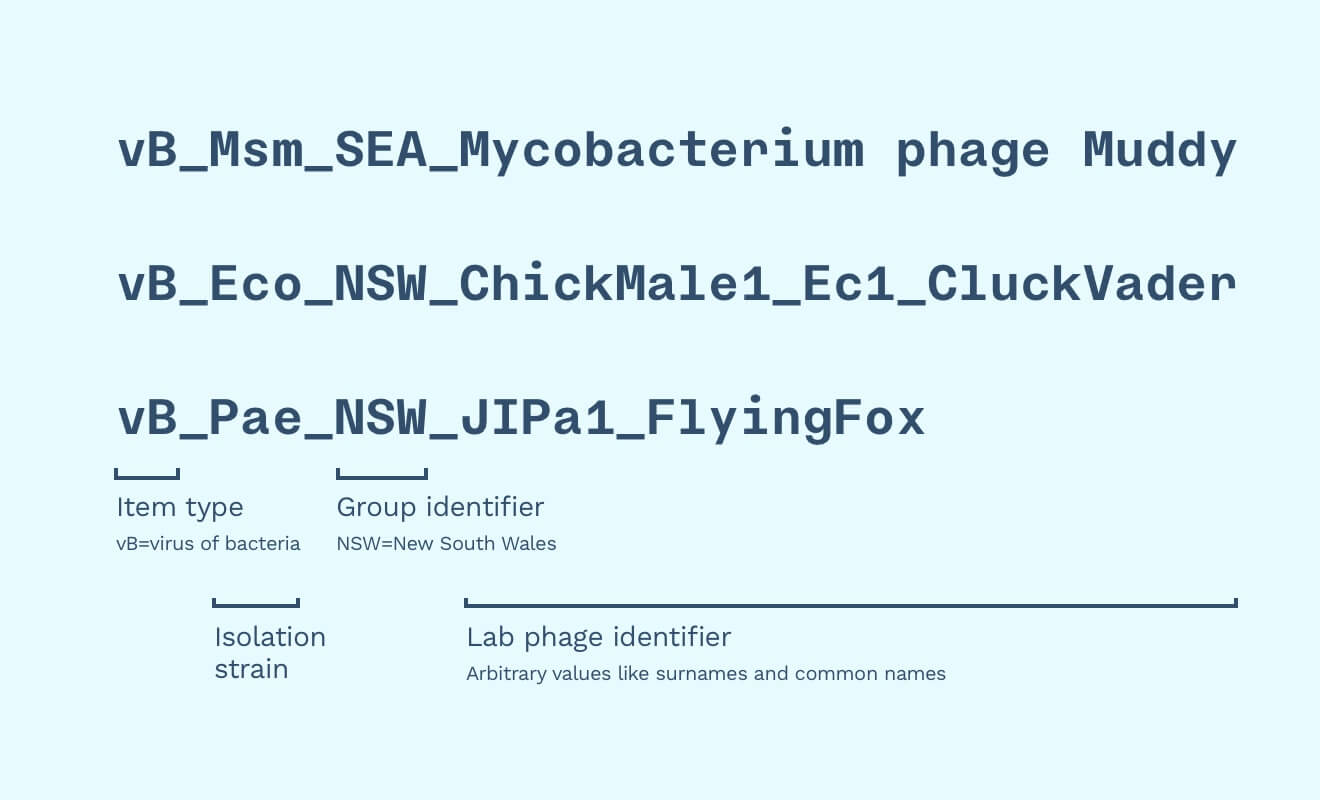
Some name example names could be “vB_Eco_NSW_ChickMale1_Ec1_CluckVader” and “vB_Pae_NSW_JIPa1_FlyingFox”. In this example, NSW stands for where the phage was isolated (”New South Wales”) and “JIPa1” stands for “Jon Iredell lab, P. aeruginosa phage number 1”, which is the first P. aeruginosa phage isolated in the lab.
Though more flexible, this naming system is complicated, pedantic, and obviously impossible to pronounce. This extra information adds a lot more contextual information and makes the phage names more easily machine readable. This in turn will make it easier to write code that can search, filter, and display phages in interfaces, apps, and reports.
When we add dozens or hundreds of phages from other collections, this extra flexibility allows us to accession others’ phages into our own collection’s identification system — without confusing where they came from. The “lab phage identifier” section should accommodate any other labs’ naming systems.
A more pedantic accessioning / naming system that includes provenance helps us track down the provenance (where a phage has come from) more easily. It helps us identify phages in our own biobank vs. other biobanks. If genetic drift or other issues occur, this naming system helps us indicate that a change in the phage has occurred in our biobank. This complicated naming system is primarily useful within the database and freezer system. Whether authors will want to publish using the accessioned database ID or its common name is purely up to the authors.
Internally, we’d still refer to our phages by its lab identifier (Pa1) or common name (FlyingFox)!
Different phages of the same name
With any kind of arbitrary naming system, sooner or later you will end up with problems like name collisions (the same name points to two different phages). This is ok, as long as you’re able to “resolve” the name of the phage to an actual box in the freezer, or row in a spreadsheet or database (e.g. if you know where the phage came from). This is why it’s very important to add a “surname” or a “group identifier” to a phage name, to help us trace the provenance. We don’t want too many “John Doe” phages floating around biobanks and publications.
At Phage Australia (and as Phage Directory) we’re working on a simple phage name registration/idenfification system, similar to the DOI system for papers or clinicaltrials.gov for clinical trials.
Naming rules can get complicated very quickly. Evelien from ICTV actively discourages adding too many characters to the name, like dashes and underscores. Ideally, she says, names should be as simple as possible.
Sequence-accurate identifiers
If we need a lot more precision around naming a phage, we could consider a phage’s characteristics in relation to its genetic and conditional characteristics (e.g. host range or plaque morphology), its abilities to lyse, or its sequencing information. If a phage is engineered, mutated, or experiences genetic drift — at what point does it become a different phage? At what point does it matter that we identify it as a different phage? E.g. do we consider a mutant “different” once it plaques on different hosts? At this point, do we give it a new name?
We might identify a phage purely by its genome sequence. This allows us to fully disconnect its identity with its characteristics and abilities. Full sequences are very hard to remember as names though — This is where hashing algorithms come in.
For example, let’s take the T4 phage genome. We can hash the contents of its fasta file using an algorithm like sha256 and turn the entire sequence into a slightly shorter line of numbers and letters. Here’s an example of what a hashed ID of a T4 fasta file looks like: “a4792ca503e4b70ff194fb981606a137dd73b43589329ca7612d36c02f08f9eb”.
Even though the hash is long, and only readable by machines, it still fits into a spreadsheet or database entry better than an entire Fasta file.
If we want to go further, we could use a web3 tool like IPFS (https://ipfs.io/) to turn the T4’s genome into a URL. This URL acts both as an ID to the genome and as a link to the fasta file itself. This is really useful for building something like Genbank:
https://dweb.link/ipfs/bafkreifepewkka7ew4h7dfh3talanijx3vz3inmjgkokoyjng3ac6chz5m
Sometimes we might have a collection of phages that are all labelled as T4, but whose genome sequences appear slightly differently from each other (e.g. because of drift, mutations, or sequencing errors). When hashed, these sequences would all have a different values — because their genome sequences all differ. Though their sequences differ, we might not want to classify them as uniquely different.
We then compare the sequences with each other and analyze their sequences’ hamming or Levenshtein distances or ANI (average nucleotide identity). Basically, this is a number that tells us how different genomes are from each other. Using this number, we can say something like “every phage sequence that has less than 3% sequence difference should be classified as the same phage” — we could set our own arbitrary cutoff numbers depending on our own needs.
If we later find out that one of those mutants exhibits different characteristics (e.g. different host range) from the others, we might give it a new identity, as it’s now functionally different from its close relatives. We might even give it a new common name!
Combining hashing and hamming/Levenshtein/ANI, gives us a new way to uniquely identify phages and their relationships to each other. As our phages undergo mutations, we end up with phages that appear closer or farther from each other (genomically or otherwise). We could represent these in “graphs”, similarly to how we currently represent family trees (e.g. as weighted, directed graphs)
I am known by many names…
A name is only useful when the majority agrees to refer to it by that name. Some phages that have moved between labs/collections will be called different names throughout publications.
Make sure to have spreadsheet that connects the phages’ common names, internal database IDs, Genbank accession IDs, and any names the phage has previously been known. Make sure it’s up-to-date when new phages are added!
Here’s a handy naming guide from SEAPHAGES: https://phagesdb.org/namerules/. And have fun naming your phages!
Looking ahead
Up to now we’ve explored identity and naming systems for our phages. Next, we can think of our phages like items on a shelf, books in a library, or items in a museum. Like inventories in a store. How do we think about, and manage our phage inventory? How do we keep track of “provenance” and “governance” and properly set up logistics?
We’ll talk through these somewhat dry, yet absolutely vital aspects and biobank requirements in the next issue.
Special thanks to Jessica Sacher, Evelien Adriaenssens, and the Phage Australia team (Ruby Lin, Nouri Ben Zakour, Stephanie Lynch, Jon Iredell) for helping me hash some of these ideas out.
More special thanks to various phage labs and biobanks we’ve spoken to over the years about data management. Some of these labs include: Queen Astrid Military Hospital, Sciensano, the Félix d’Hérelle Reference Center for Bacterial Viruses, DSMZ, ATCC, NCTC, TAILOR, Israeli Phage Bank, The Bacteriophage Bank of Korea, Fagenbank, Citizen Phage Library, Japan Phage Bank, and many more, throughout the years. Thanks so much for putting up with my incessant questioning!
Further Reading










