Hi everyone, it’s me again!
This week I’ve been exploring ChatGPT’s various capabilities like data analysis, image upload, optical character recognition (OCR), and more. I’ll walk through some of my experiments, that surprised me along the way.
Before we get started, this post uses the paid GPT-4 version, and the updated “GPTs” upgrade that came out a couple of weeks ago. Many of the ChatGPT transcripts I’ve shared require a subscription, but I’ve also attached many screenshots for everyone to view.
If you’d like a refresher, read our last post before continuing: **ChatGPT tips for lab work.
Exploring ChatGPT’s capabilities for data engineering
I like to use ChatGPT for processing a lot of data, or “data engineering.”
Data engineers make sure data is collected from various sources (like the clinical lab, the wet lab, the genomics side) into a single place that’s easy for other people to use. Data scientists analyze, interpret, and make sense of that data — usually by creating models, predictions, and help make informed decisions.
Usually, this is a lot of work, and many organizations have huge teams of data engineers and data scientists — and lots of complex and expensive tools and software. Rote data tasks like correcting spellings, combining duplicate rows, and fixing typos required either manual tasks or complex/expensive software. The worst part of data science is “cleaning” the data.
Another tough problem is “extracting” data. This includes grabbing text from printed or handwritten text, turning graphs and tables to structured data, or even counting plaques on a plate.
In the following experiments, I’m throwing GPT-4 some familiar workflows to see if it’s up for the task!
Extracting data from raw text
I often need to turn images into structured data, which I do by hand (very time consuming). With ChatGPT, I was able to grab a screenshot of the TAILOR’s HP3 phage’s Physician Desk Reference card and turn it into structured text.
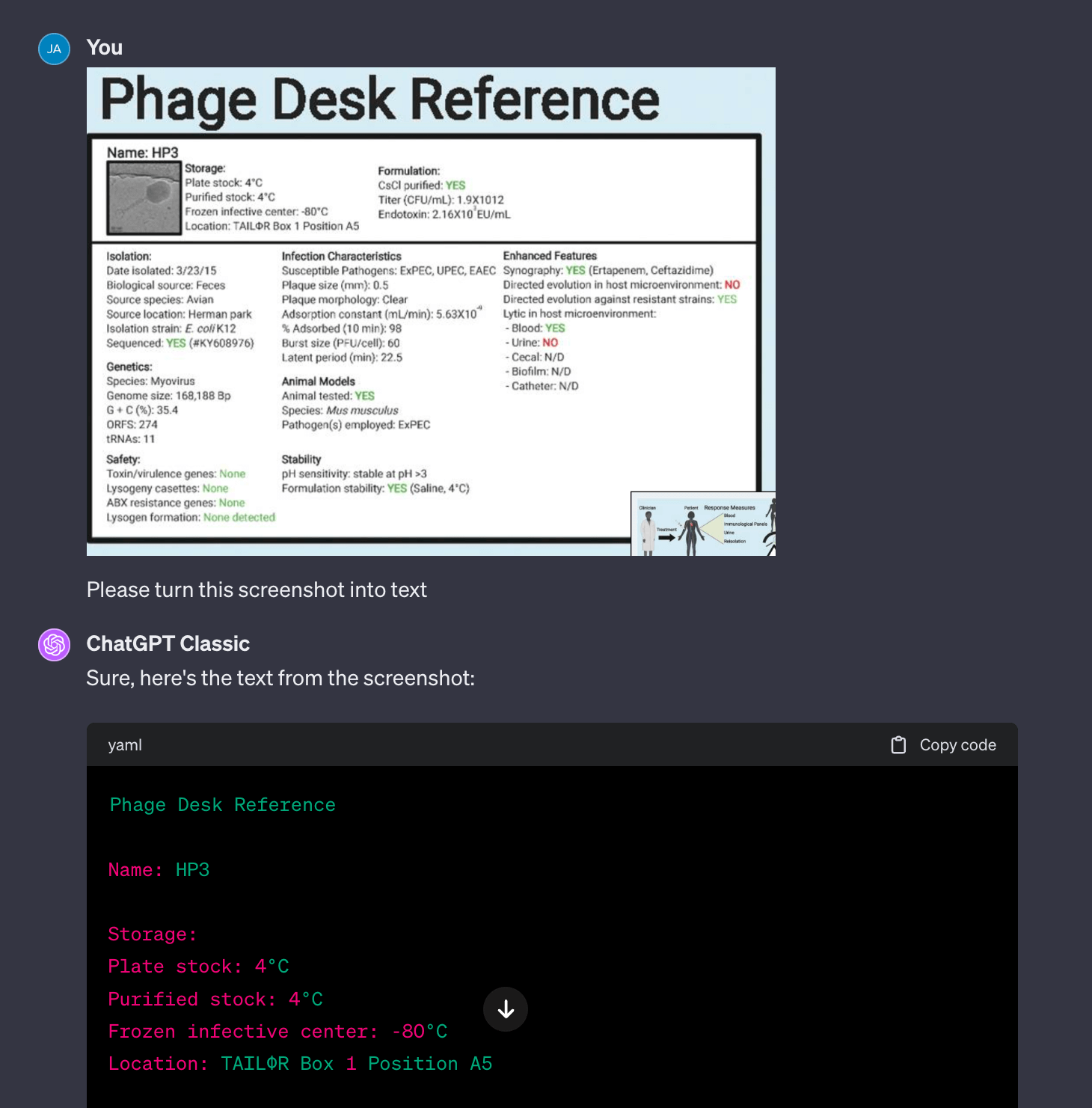
Fig 1. Extracting text from TAILOR’s Physician Desk Reference example from their paper is a simple task for GPT-4 (link to full image)
One annoyance with the default GPT-4 is that it prefers to run Python to extract the text (which it does poorly and fails over and over). What works much better is switching to “Classic ChatGPT” which turns off the ability to run Python. This version uses the multimodal (the model’s inherent text and image capabilities) system to extract text from the image. As you can see from the example, it’s properly transcribing and structuring the text!
Turning screenshots into structured data
GPT-4 works with text, but what about graphs and charts? Lots of paper report data as graphs and charts, but don’t back it up with actual tables or CSV files. To get any kind of analyzable data, traditionally you’d have to manually copy every line into Excel. How does GPT-4 fare?
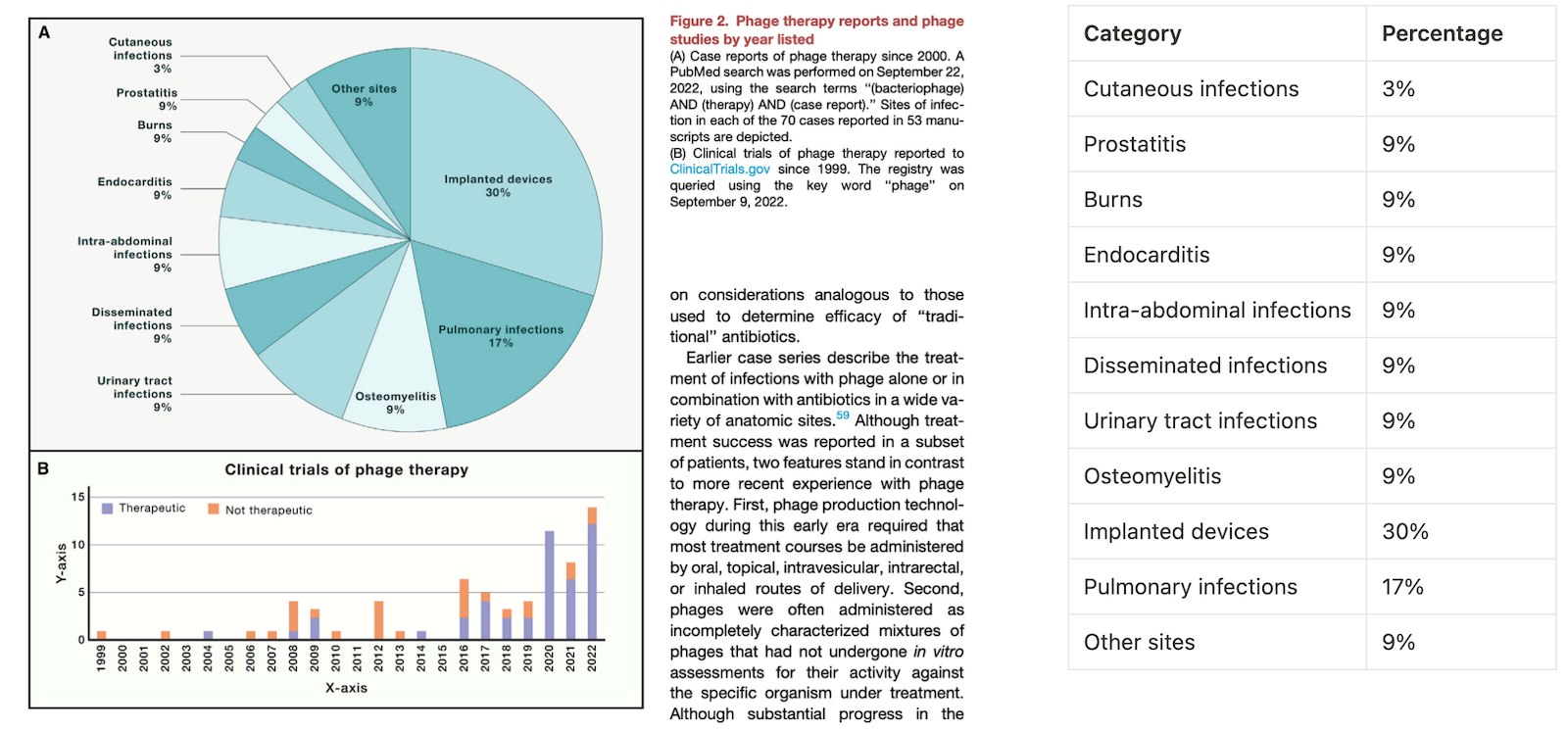
Fig 2. ChatGPT is easily capable of extracting Chart A into a table (right), but struggled to extract Graph B. (Transcript from failed Chart B extractions) Source: Phage therapy: From biological mechanisms to future directions by Steffanie Strathdee, and others
It seems that ChatGPT has no problem extracting the data from the Chart A (the pie chart) as the labels and values are spelled out. I added the extracted information on the right.
However both the default GPT-4 and Classic ChatGPT continuously struggles to translate graph B into a data table. While the default GPT-4 is unable to use a Python module to extract the data, classic ChatGPT keeps making up fake numbers. Though the data is not labeled, this should be an easy task for humans to estimate, but GPT isn’t able to guesstimate the values around the correct ballpark.
I also tried to extract data from the KlebPhaCol collection’s host range. It really struggled to map each cell to each strain range — either making up values or stating it’s unable to — which really shows its limitations in extracting accurate data from rich charts and graphics.
Though GPT-4 is great for explaining gist of images, it shouldn’t be relied on for pure data extraction (unless all the values are clearly labeled with text).
Explain a graphic
From the same paper as Fig 2, I asked GPT-4 to explain a graphic on phage engineering. I conveniently left out the caption, to see how close GPT-4 would get. Surprisingly, GPT-4 did a great job capturing each panel. For a non-biologist like me, this tool is invaluable to help me build a basic understanding of papers (without having to bother Jess every five minutes).
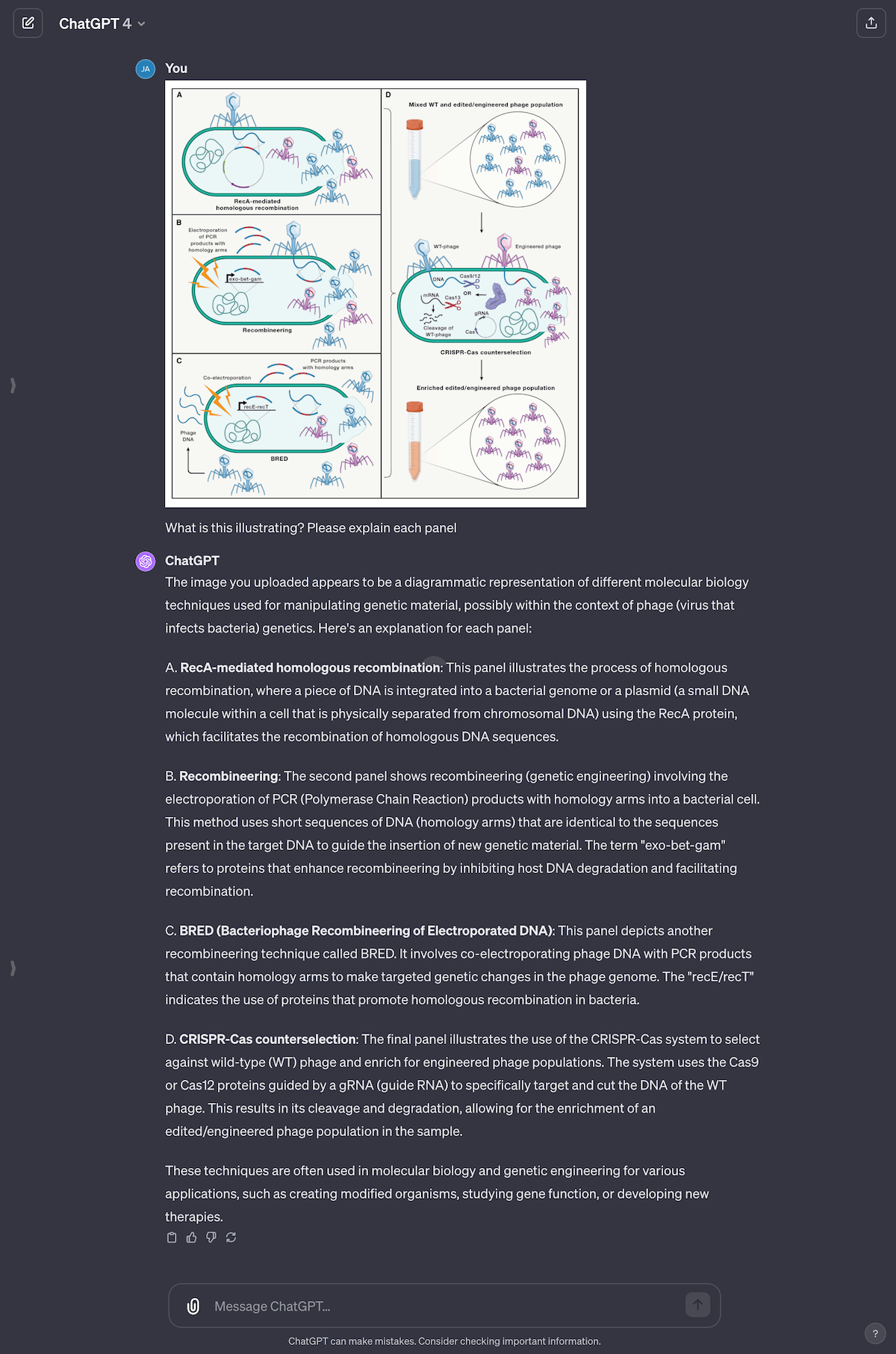
Fig 3. I asked ChatGPT to explain this image, without giving it the image caption. Looks it did a great job! It’s hard to know if it got that from reading the text, or from the graphic itself. (Full image) Source: Phage therapy: From biological mechanisms to future directions by Steffanie Strathdee, and others
Processing QR codes and bar codes
I thought processing QR and bar codes would be straightforward, since it’s a solved problem, but it’s not. It looks like ChatGPT has an old module of cv2 which for some reason can only read QR codes but not bar codes (even though bar codes have been around for way longer). ChatGPT also seemed to struggle to analyze the image and generate an answer, even for the QR code that worked. This might because of the new GPTs architecture they’ve moved to, and will hopefully improve over time.
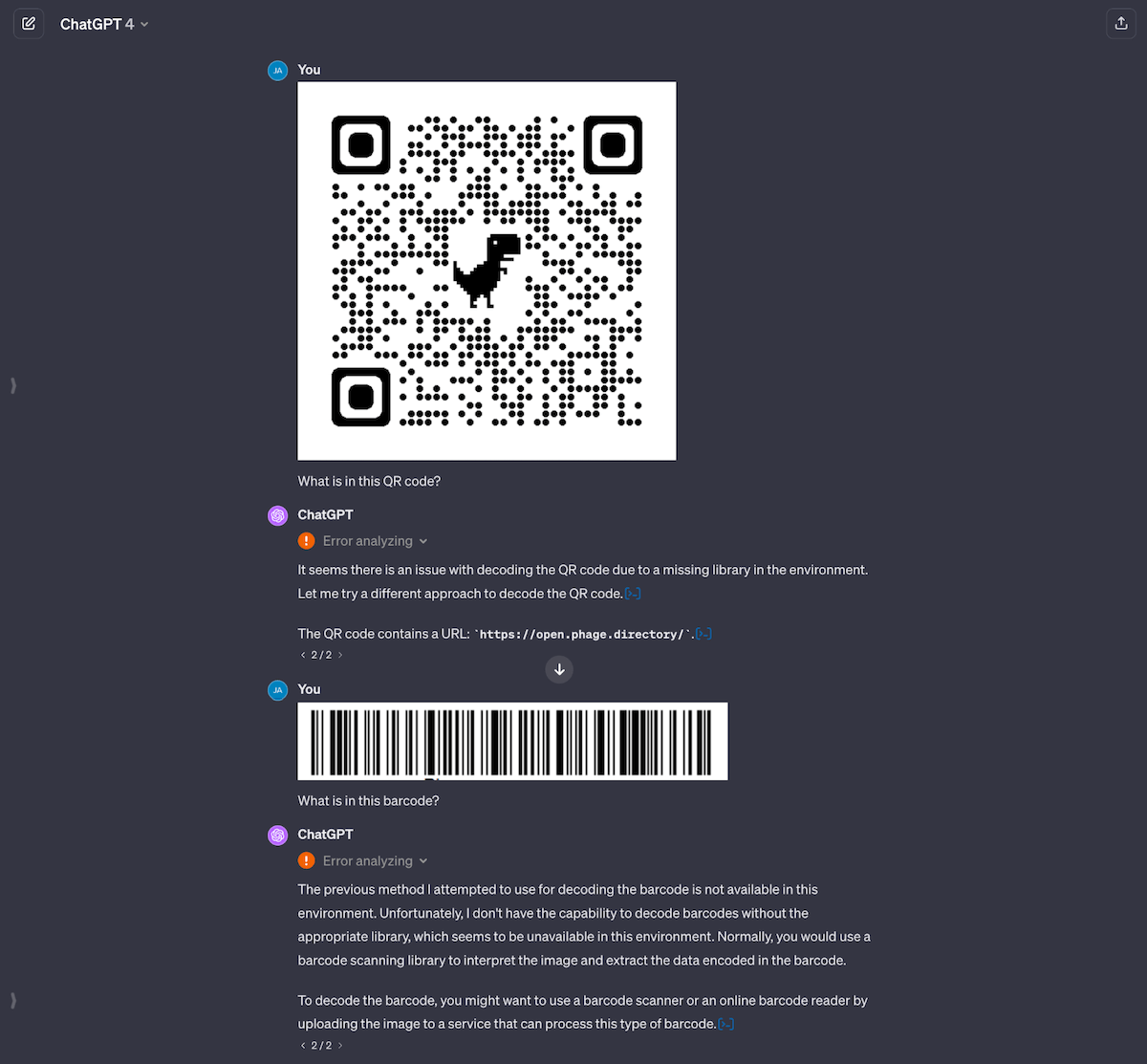
Fig 4. GPT-4 is able to read the QR code (after two tries and some prodding), but is completely unable to read the bar code. Under the hood, GPT-4 analyzes the image using Python’s cv2 module to return an answer. In the case of bar codes, its module doesn’t seem to support bar codes.
The inability to read bar codes is actually the biggest surprise to me, as doing that is trivial in Python and should only take a couple of lines of code. This convinces me that ChatGPT itself wouldn’t suffice as a production pipeline, as you can’t control what tools it has access to underneath the chat app.
If I built a custom chat system, I’d be able to provide tools that I knew my team needed. When using the standard ChatGPT, we’re at the mercy of what their developers decided to add to the system. However, custom GPTs are always an option, and I’ll cover how to build your own in an upcoming issue.
Turning handwritten notes to Markdown
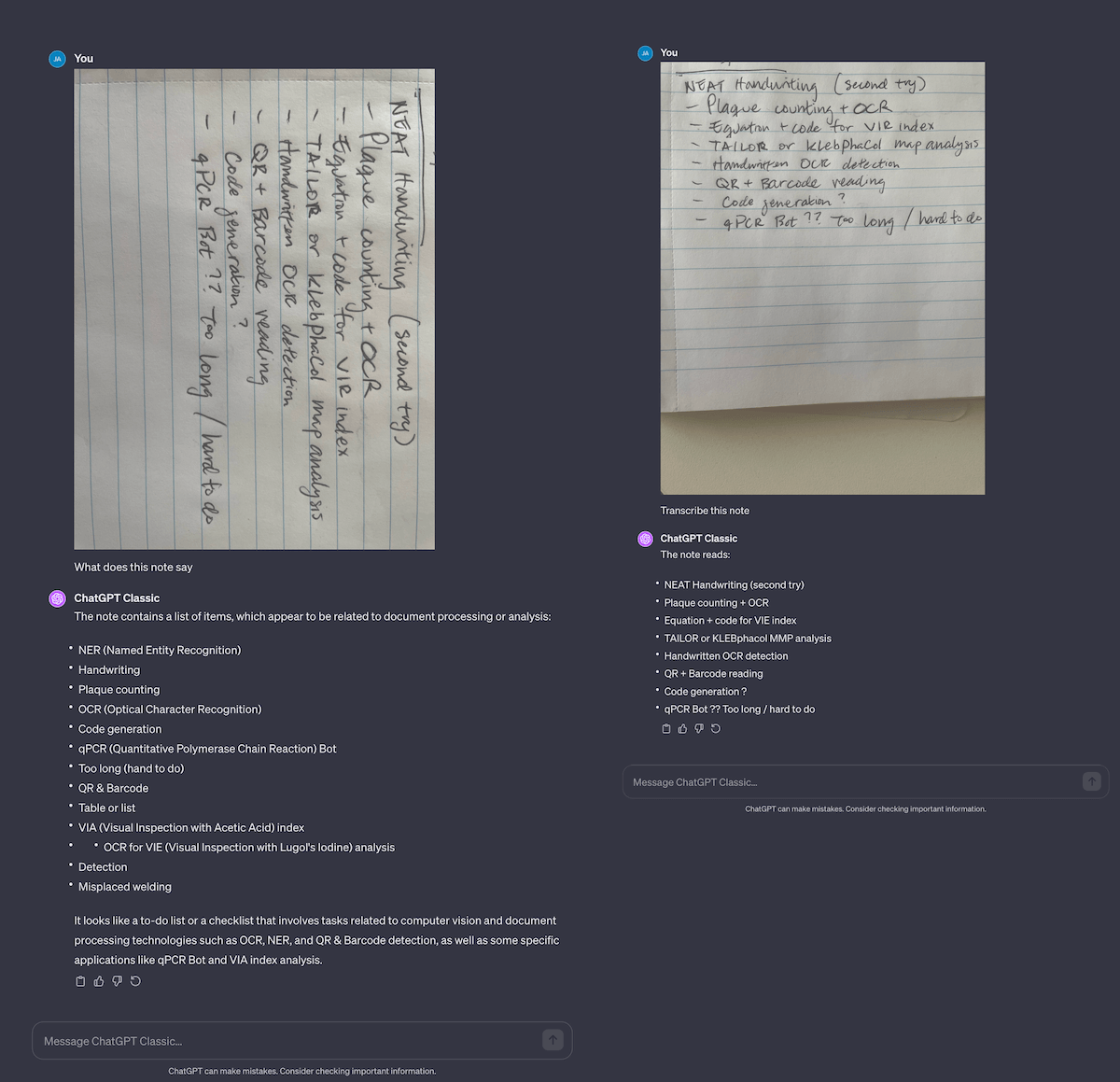
Fig 5. Comparison of extracting handwritten text. My original note… didn’t work very well, so I had to clean up my handwriting. This worked surprisingly well, and convinces me I should probably start writing neater! Notice the mayhem that ensues when using an incorrectly-oriented picture…
Another massive task is turning handwritten lab notes to digital notes. This is a difficult task for humans, let alone computers. Fo example, I have trouble reading and transcribing Jess’ lab notes; from shorthand to hard-to-read values. I tried to do this on my notes, and it fails fairly hard… until I rewrote them with neater handwriting! It seems to get 99% of the text correctly (which still means 1 of every 100 letters is wrong), but it’s easier for me to go back in and fix, rather than type everything from scratch.
Curiously, if you give it a note in the wrong orientation, it will wildly hallucinate answers! If you’re automating OCR with GPT-4’s API, make sure you detect for image orientation and rotate it correctly before sending the image to the API. Otherwise you’ll have a mess on your hands.
Counting plaques with ChatGPT
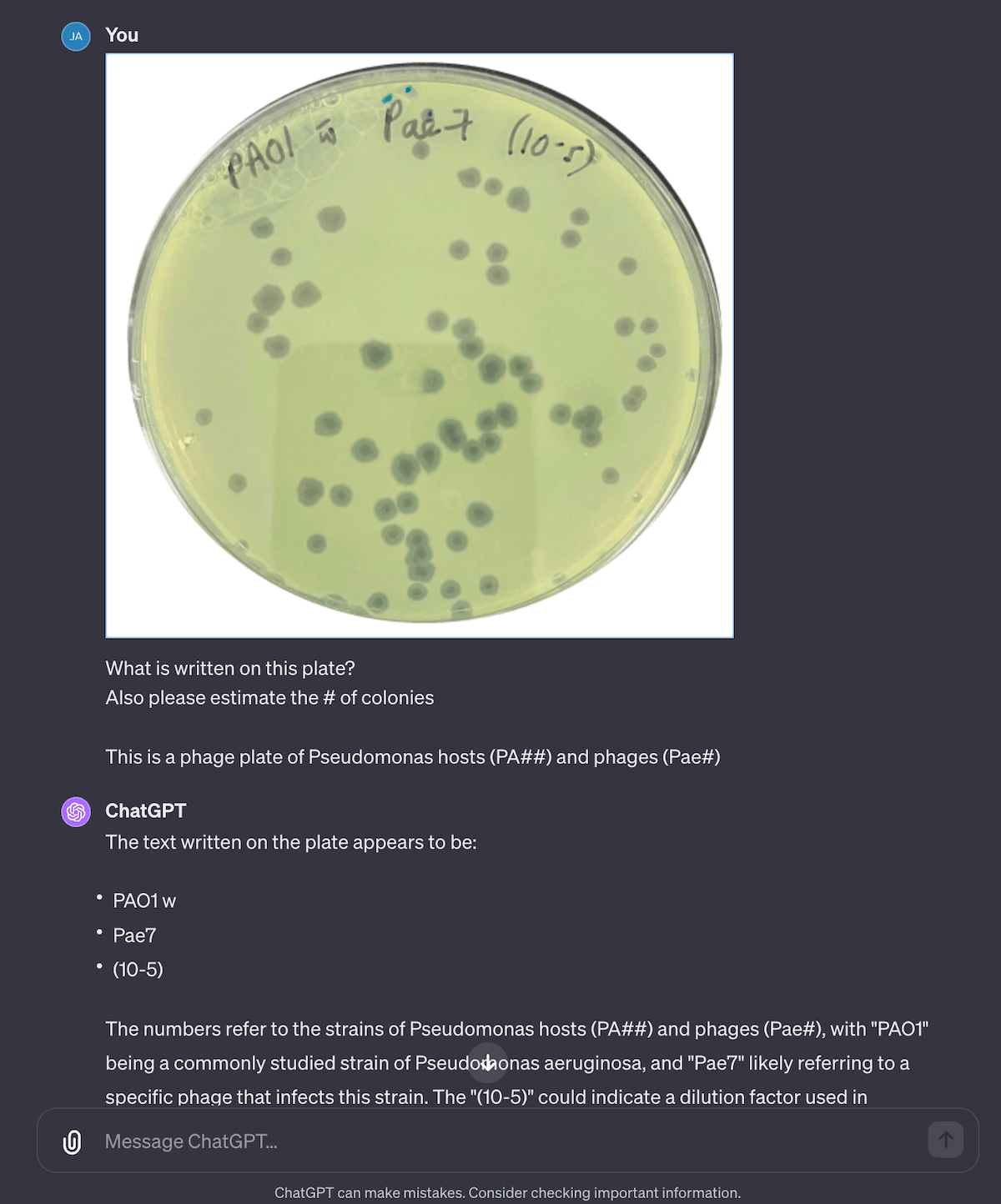
Fig 6. Counting plaques is hit or miss, but it’s surprisingly good at reading plate Sharpie writing! Especially when you guide it, by saying how the lab labels hosts and labels. Generally feeding it a “schema” on the correct keywords, labels and nouns makes it much more correct.
This was a task I didn’t think it could complete… and it kind of worked. I uploaded one of our old plates for PhageAustralia’s Pae7 and it did surprisingly well. By telling it , it’s able to read the text on the plates fairly well. When it comes to plaque counting though, it just makes up numbers, and is very inconsistent, ranging from ~60 (which seems accurate) to 66,000+ (…not so accurate). Most of the time time the answer falls within the ~50-100 range (I ran it 8 times) but it’s still way too off to be trusted. I’d try connecting it to Plaque Size Tool or OnePetri as an external API action.
Analyzing FASTA files?
Unfortunately, ChatGPT won’t become your AI bioinformatician any time soon. The standard GPT-4 implementation doesn’t have any capacity to run any bioinformatics tools and workflows like Nextflow or Snakemake, and the standard environment is also missing tools like Biopython (transcript). Apparently you can upload Python code / “wheels” and it’s able to run those, but I haven’t tried that yet. I think with a custom GPT connected to either your own cluster, server, or a cloud-based tool like Google Cloud Platform or latch.bio, you could potentially get a GPT to run bioinformatics workflows. Keep in mind that GPT tends to get a lot of stuff wrong, so you might be running many workflows that just don’t work.
Using ChatGPT for repetitive work
In this section, I tried to use ChatGPT for various tasks I’d normally have to do manually. Generally the outcomes were pretty good, and I shaved many hours of work, but the outputs aren’t perfect. Though admit, it probably did a better job than I could ever do.
Collecting bioinformatics resources by combining documents
Last week I launched the Open Phage Data Sheet project, an publicly-editable Google Sheet of phage resources. However, many tabs were mostly empty! I wanted to write about the process of using automation, code, and AI to fill those sheets.
I’ve now filled the bioinformatics sheet — which is now around ~400 tools — with the help of ChatGPT. I’ve started by merging Phage Kitchen (our bioinformatician Nouri)’s resource and Rob Edwards’ Viral Bioinformatics Tools spreadsheet.
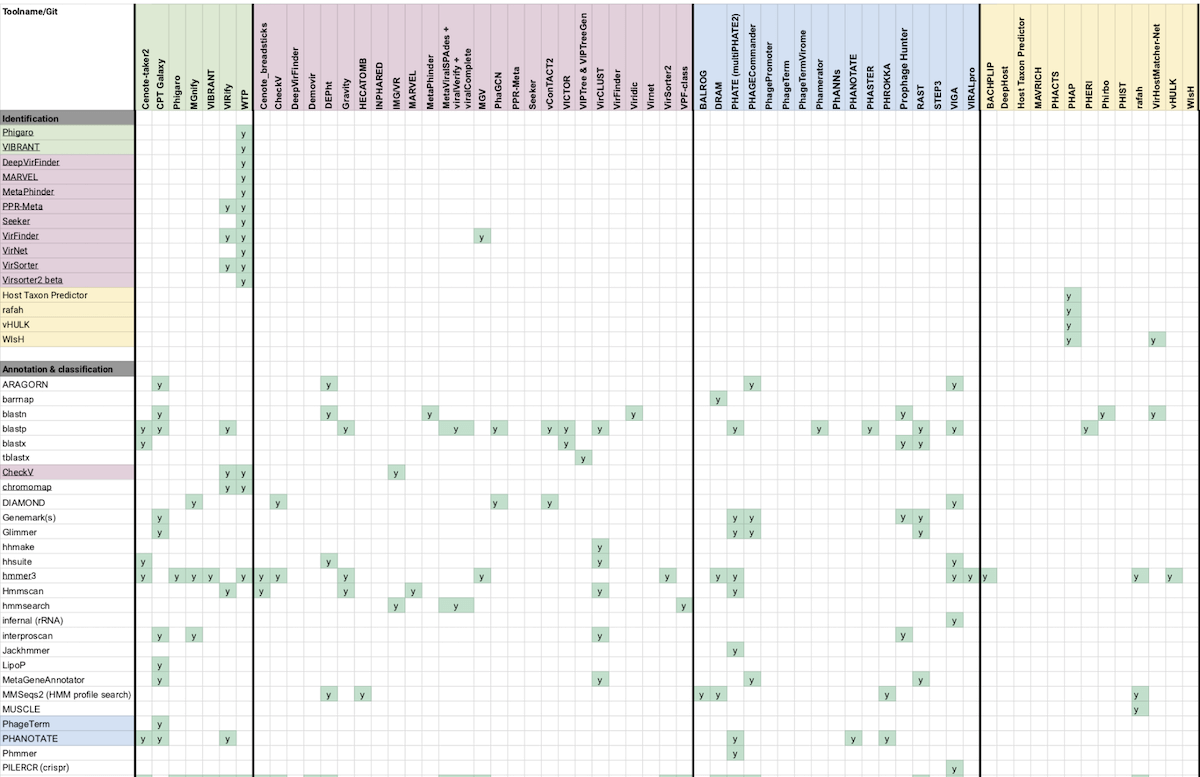
Fig 7: Merging Phage Kitchen’s color-coded, categorized list of bioinformatics tools and compatibilities would normally take a bit of manual work, but GPT-4 seemed to accurately extract each tool and type into my sheet.
Phage Kitchen is a Github repository of a few distinct resources: a spreadsheet of compatible tools in different categories identified with a color coded legend, a Trello board of tools with supplementary descriptions and screenshots, and a few markdown documents of tools broken into categories like Complete pipelines and Image analysis.
Rob Edwards’ spreadsheet is laid out in rows and columns, but has an “other lists” category that’s more like a document of notes and tools.
Transforming and merging the data
The first thing I have to do is somehow bring all the names, descriptions, URLs, and categories of tools from all these various places into my own spreadsheet, according to my own columns (schema).
Traditionally, merging all this information is really tough. I’d have to write scripts to transform Rob’s sheet to match my own spreadsheet. I’d have to go through Nouri’s color legend, add a column, and add that category to each tool. I’d have to figure out how to move Nouri’s Trello and markdown docs to my sheet. Thankfully, OpenAI’s Data Analysis tool makes things a bit easier!
The first thing I did was copy/paste Rob’s Bioinformatics “other lists” to GPT-4, and asked it to create a table based on the columns: Name, Category, Notes, Author, URL, Citation. Then, I tried copying the entire list of 162 tools, which really tripped it up (transcript). ChatGPT kept taking forever and crashing multiple times, and eventually I gave up. This might have to do with their scaling issues, and why they’ve momentarily turned off new signups.
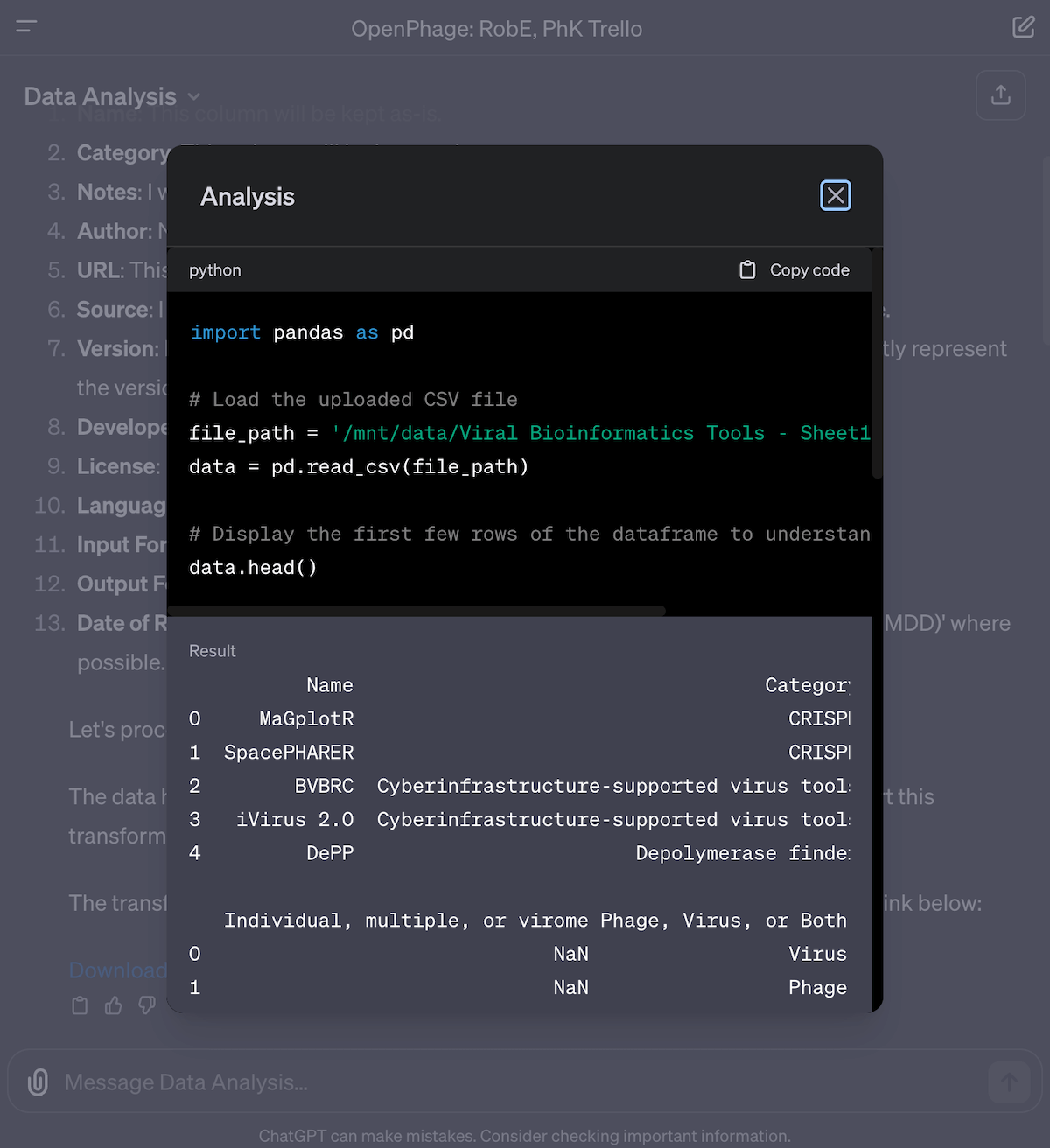
Fig 8. The Data Analysis GPT writes and executes python code on your data. You can actually copy this code and reuse it later by running it on your own machine!
To make it work, I had to switch to the Data Analysis GPT (previously called “Code Interpreter”). This tool analyzes the spreadsheet, writes out the steps to process the data according to my wishes, then writes and executes Python code to create the outputs. You can even click to see the code it’s written under the hood, and copy it for yourself. With Data Interpreter, I uploaded the Viral Bioinformatics Tools CSV, downloaded the JSON file from Trello and gave that to ChatGPT, and uploaded the separate descriptions as Markdown files and the Google Sheet as an Excel file (so all sheets would be uploaded).
When using the Data Analysis GPT, don’t upload all the various files at once and ask it to merge all the data — it’ll get confused or fail outright. Instead, I ask it to process one file after another, while repeating the instructions. Every file I uploaded, I asked it to merge that dataset into the main Bioinformatics list. This reduces errors and speeds up the process.
After I’ve asked it to merge all the data, I then ask it to clean up and merge duplicates, and then save the final result into a CSV. I then download the CSV, open it up in Excel, and copy the results back to the Open Phage bioinformatics sheet.
With this process, I’ve been able to merge several sheets into my own sheet, which now has ~400ish tools! What’s really neat is that things that were previously very annoying to do — like extracting links from various text sources and moving them to an associated column, deduplication rows and merging their values, converting color-coded data to structured data — all this would have taken at least a day or two for me to figure out. All the work listed here probably took 2-3 hours. Most of that time was spent on finding links, figuring out what to combine, waiting for ChatGPT and manually verifying the results by spot checking.
Here’s exactly what I did, along with shared ChatGPT transcripts (requires the paid version of ChatGPT).
- Upload the Viral Bioinfo sheet as CSV, and the Trello JSON (ChatGPT)
- Merge Phage Kitchen’s Excel spreadsheet of tools (ChatGPT)
- Add Phage Kitchen’s descriptions of tools (ChatGPT)
- Fix duplicates and do a light cleanup (ChatGPT)
- For verification, I only did some light spot checking. There are some errors with formatting and duplication, but I decided to do that work in the future.
Unfortunately, AI is great but it didn’t do ALL the work for me. There’s still more work to be done — like removing more duplicates, add and clean up links and notes, perform a search on each tool to get the latest link and project status, and so on. But I’ve still saved many many hours of work already (without ChatGPT, I probably wouldn’t start this project in the first place), so there’s still many benefits to using it. Take a look at the sheet: https://open.phage.directory/sheet
Help me understand a paper
The last experiment I tried was using ChatGPT to understand a paper. You can easily upload the PDF of a paper, and it’ll break it down for you. I asked it to summarize Dom Sauvageau’s Virulence Index paper in the following example, which it did a great job of doing.
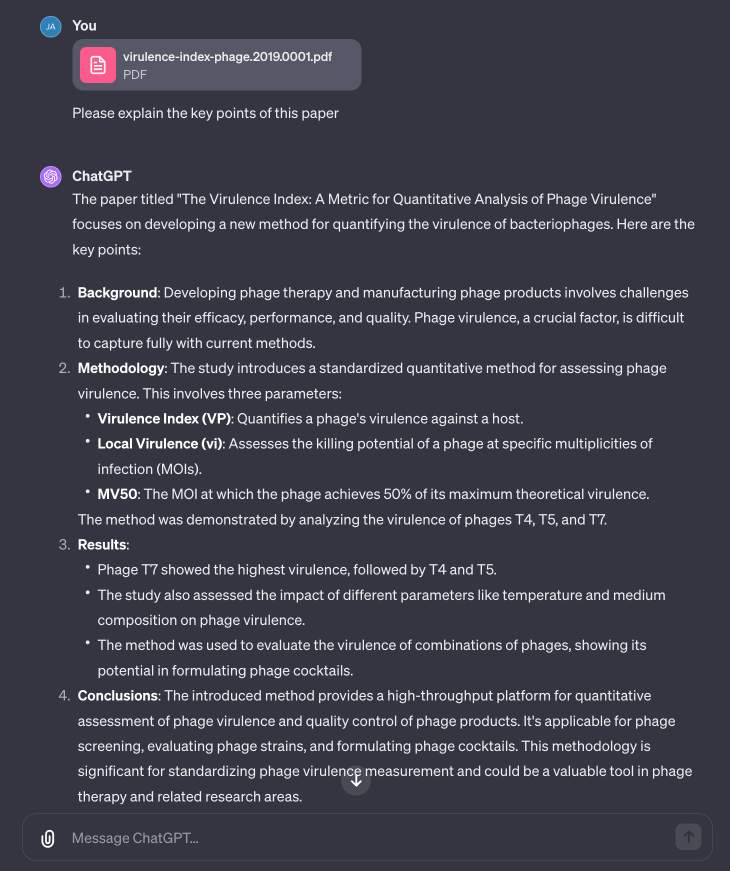
Fig 9. ChatGPT is great at summarizing or answering questions about uploaded papers. Note that for really long documents over 18k characters, others have noted that its Q&A quality tends to go down for the middle parts of the document.
I wanted to understand more about the entire process, from “how do you calculate the virulence index” down to “what is local virulence” to “how do you even calculate MOI” to get a better understanding of the entire process. I then tried to get ChatGPT to generate all the code for me to build a local virulence and virulence index calculator as a web service… and it did it for me!
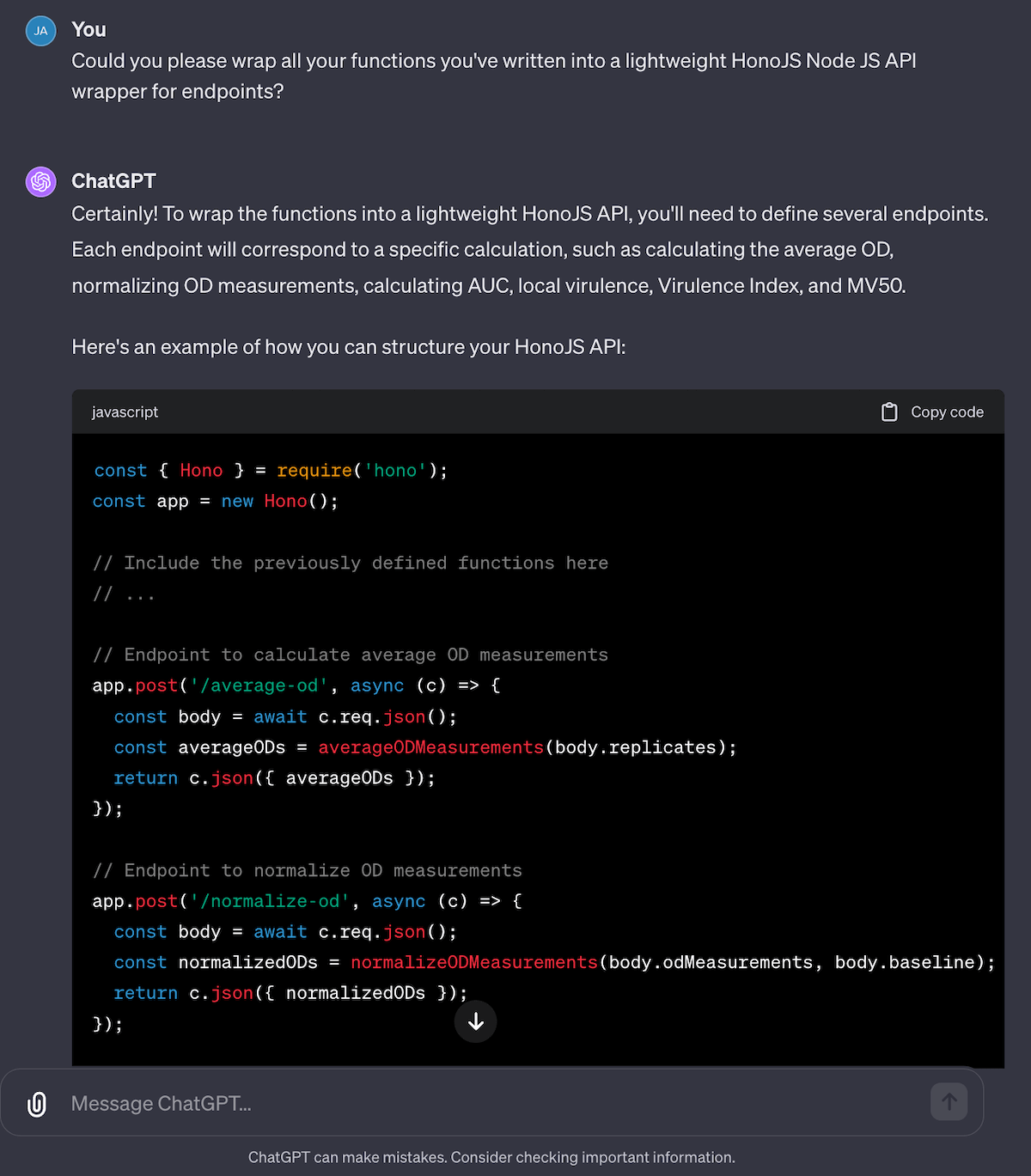
Fig 10. Not only can it summarize papers, explain equations and things like “MOI” to me; it can even take the ideas from a paper and turn it into a web application by writing all the code!
I haven’t built and deployed this yet, but this is definitely something I’ll do soon. I think it’s super powerful to have a system to not only summarize a paper — but then to explain the equations, the jargon that go with it, and even the steps to take the paper from idea and equations to runnable code.
Final takeaways
All-in-all, I’ve been very impressed with ChatGPT, GPT-4, and all the new tools available to us through the new “GPTs” system. The default, “smart” GPT-4 is ever-so-frustrating, between incorrectly choosing to use Python when it should be using its own multimodality, or its penchant to crash often. I’m also surprised by how unintuitive results can be — it’s good at some tasks I think it shouldn’t be at, and vice versa. I also think the real capabilities lie in building custom GPTs connected with custom actions that can run on my own servers. This includes connecting GPTs to our own tools like Biopython or cloud bioinformatics tools like latch.bio. This also includes connecting Google Drive and Notion so we can ask questions about them, or connecting GPT-4 to a database so we can extract and save results!
Jess and I have been playing with GPT-4 for recording lab work, improving SOPs, and lots more, and we’re excited to share more demos and tools we’re building to integrate these new tools with our current practices and procedures.
In the near-future, I’ll share more of what we learn, how to build your own custom bots, and how you can supercharge your lab work with these tools.
Best,
~ Jan















