We’re excited to share part two of the results of our State of Phage 2020 Survey! For this snapshot, we’re focusing on bacterial strain collections: which bacterial strains are phage labs collecting, how many do they have of each, and where do they get them from?
Read State of Phage #1: A wide diversity of backgrounds & phage experience here
A refresher: why we did this survey
The phage field is clearly growing around the world, but we realized that no one has collected a bird’s eye view of what everyone is working on. From our conversations with researchers, we believe that phage research activity and diversity of approaches is greater than the published literature would suggest. We created the State of Phage 2020 survey to fill this gap.
We hope it will help researchers learn more about how their phage research approaches compare with others around the world, and that it will help establish a baseline for the current state of phage research so we can collectively track how it grows and evolves over time. Check out part one of the survey results, which we wrote about here if you missed it, or want a refresher. In part 1 we focused on demographics, ie. who responded to the survey? What stage of career are they at, where are they located, and what level of phage experience do they have?
In future issues, we’ll release more data, digging into the phages people are collecting, where they got them, the kind of phage work they do, the methods they use, where they like to publish, their thoughts on sharing and commercializing phages, and more!
Our plans & hopes
This issue marks part two of a series that will wade through the data, looking at a different theme or two each time, but we’ll also publish a full report at the end of it all. We hope researchers will read it, learn lots about the field, and share it with others to inspire them to join the community and pursue their own phage research (or maybe fund it!).
Some caveats before we begin
Our survey is self-reported and importantly only approximates the phage field, so please take all insights we present with a grain of salt! The data is not statistically significant, and is highly biased towards Phage Directory’s reach. With that said, we can still glean lots of insights based on what we’ve collected!
Recap: who’s responded to the survey to date?
We’re thrilled to have received 136 full responses to our survey. Filling this thing out was no small feat, so thank you all so much for taking the time to let us know about the kind of phage work you’re doing!
Now, let’s dive in!
Why ask about strains?
All phages need hosts, so all phage labs must have bacterial strains. But how diverse and how large are these collections, and where did labs get them from? We wanted to ask about strains because even though labs may not report having phages targeting a certain strain, it’s probably only a matter of time until they start finding phages against it, if they’ve got it in their lab. Put differently, if anyone’s going to have a certain type of phage, it’s probably a phage lab who already works with that type of bacteria.
In total, respondents have collected bacterial strains spanning 62 genera and 133 species
First thing’s first: we found that 94.8% (129 labs) of our survey respondents reported that they have bacterial strain collections. We were excited to see that phage labs collect a wide diversity of bacterial strains! We weren’t sure what to expect going in (for instance, do phage labs tend to work on a small number of reference strains, or are they each specializing in their own bugs?). But as you can see from the word cloud below, there’s a pretty massive diversity of strains being collected.
The data shows that traditionally-considered reference species like E. coli and Staphylococcus are most popular, which is to be expected. We see the ESKAPE pathogens jumping out as popular strains to collect too, suggesting that clinically-relevant species are commonly collected. We also see lots of plant and animal-related strains. But from there, it’s a jungle!
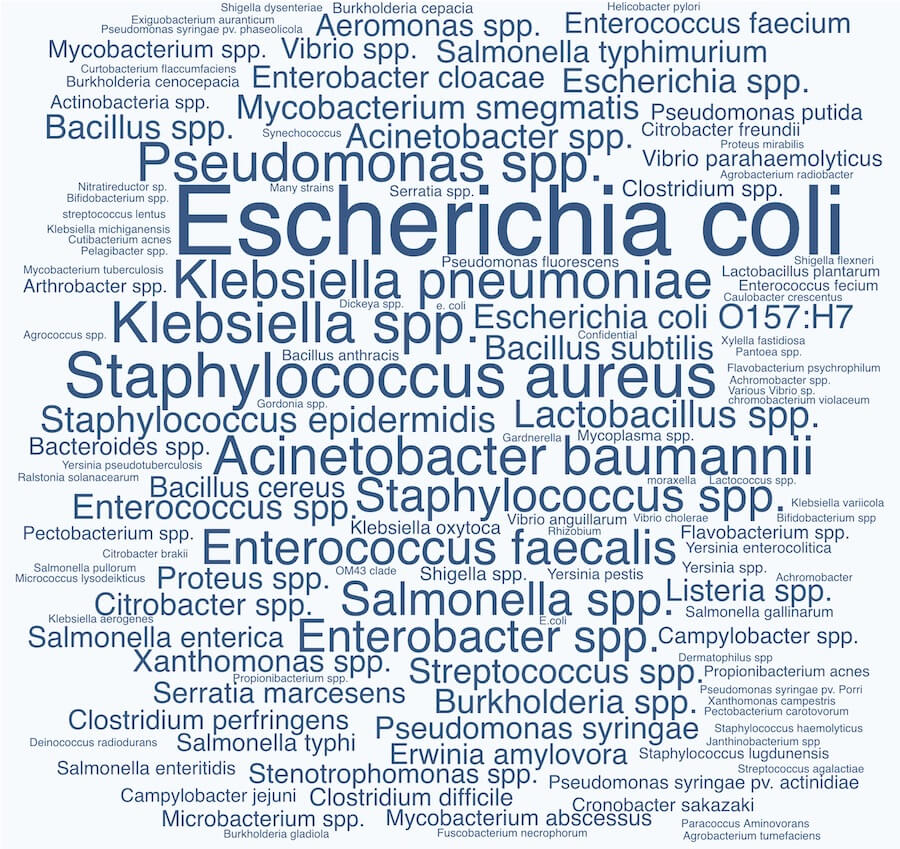
Word cloud showing the 133 bacterial species collected by phage labs who responded to our State of Phage 2020 survey. Font size is proportional to number of labs who reported that species. View larger image
Collecting a few strains per genus is most common, but some labs have large collections of strains almost no one else has
For each bacterial genus collected, we asked respondents to estimate how many strains of that genus they had (either 1-10 strains, 11-50 strains, or 50+ strains). Most labs seem to collect fewer than 10 strains of a given genus (green bars in the graph below), but a fairly large subset of labs collect more than 10 strains of a given genus (yellow bars: 11-50 strains; blue bars: 50+). Interestingly, there are a few labs that collect very large numbers of strains for a some relatively unpopular genera, including Gardnerella, Dickeya, Pectobacterium, Yersinia, Vibrio, and Flavobacterium. We also noted that Mycobacterium, although widely known from the SEA-PHAGES program to be the genus used most commonly for phage hunting, is collected by only a minority of the respondents of this survey. This shows that most of our respondents likely do not come from the SEA-PHAGES program.
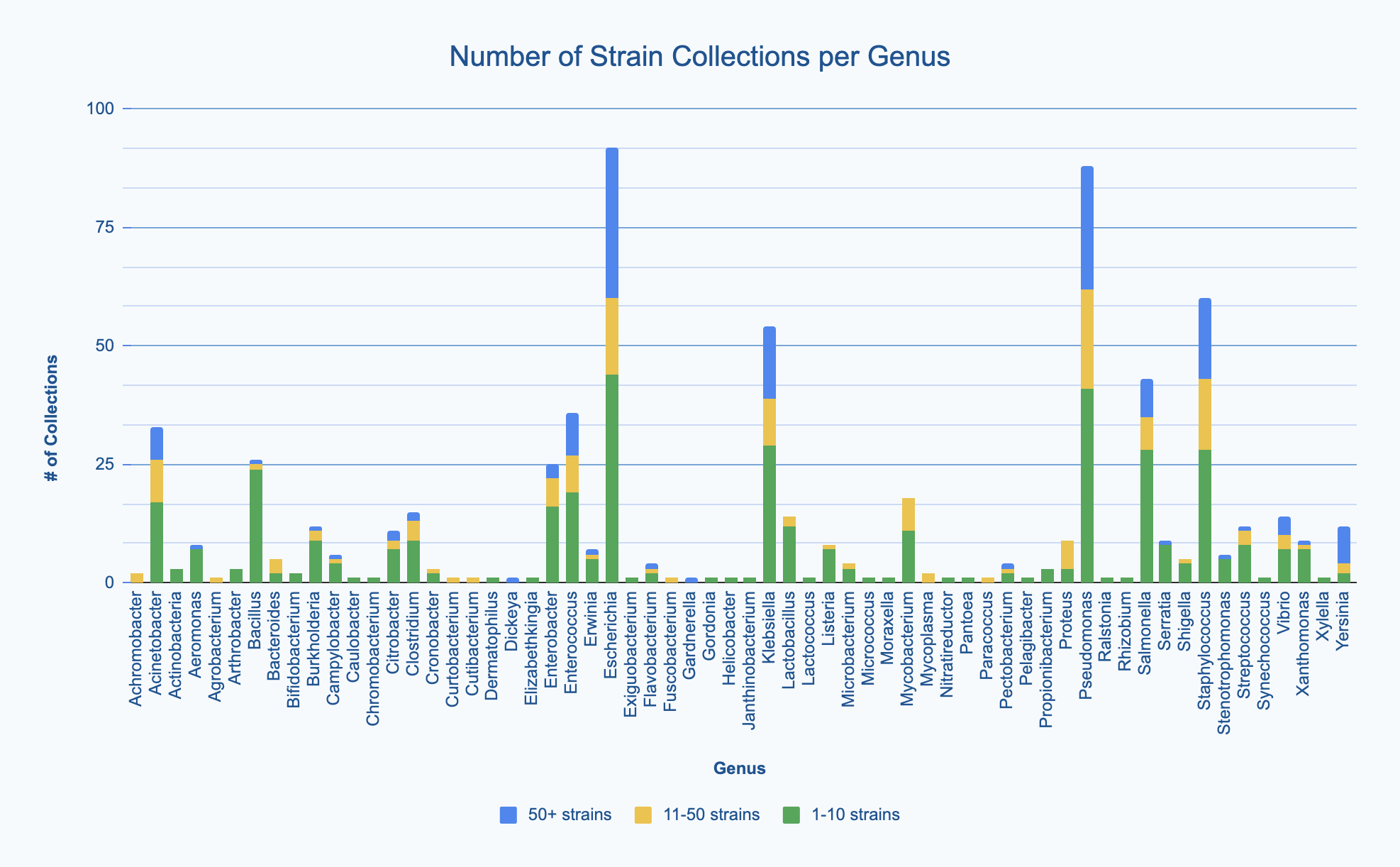
Size of strain collections for each bacterial genus collected. View larger image
Each continent collects bacterial genera in its own proportions
We wanted to know where in the world different strains were being collected, to see if we’d see any interesting trends. We divided countries into continents/regions, then looked at relative proportions of bacterial genera collected by each. Some of the things we noted were that Klebsiella is as common as Escherichia in Asia, Salmonella is very popular in Africa, Vibrio is especially popular in South America, and Staphylococcus is less commonly collected than Escherichia and Pseudomonas across every geographic area. As expected, ESKAPE strains are popular around the world. We also noted that respondents from Europe report collecting the widest diversity of genera.
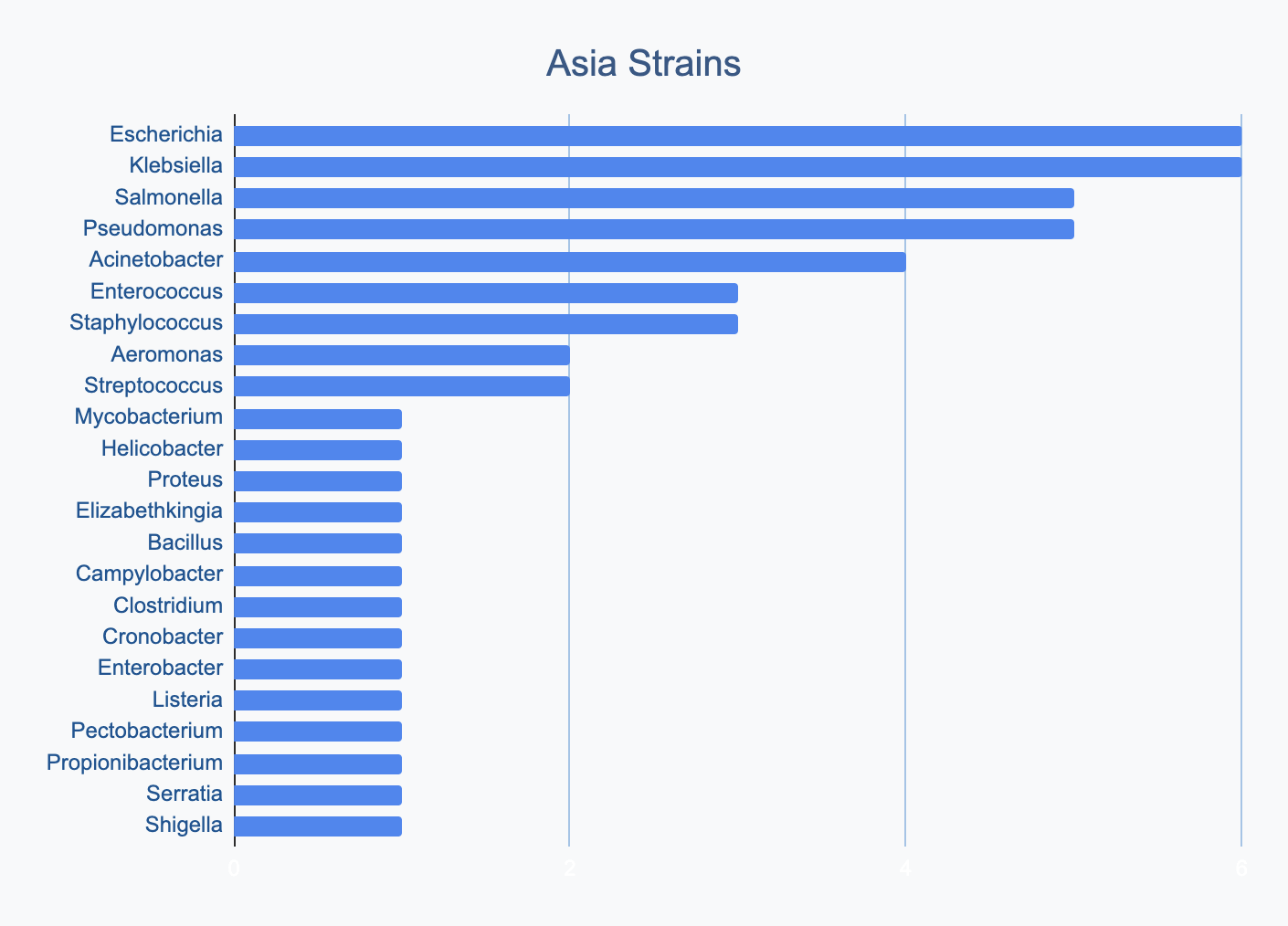
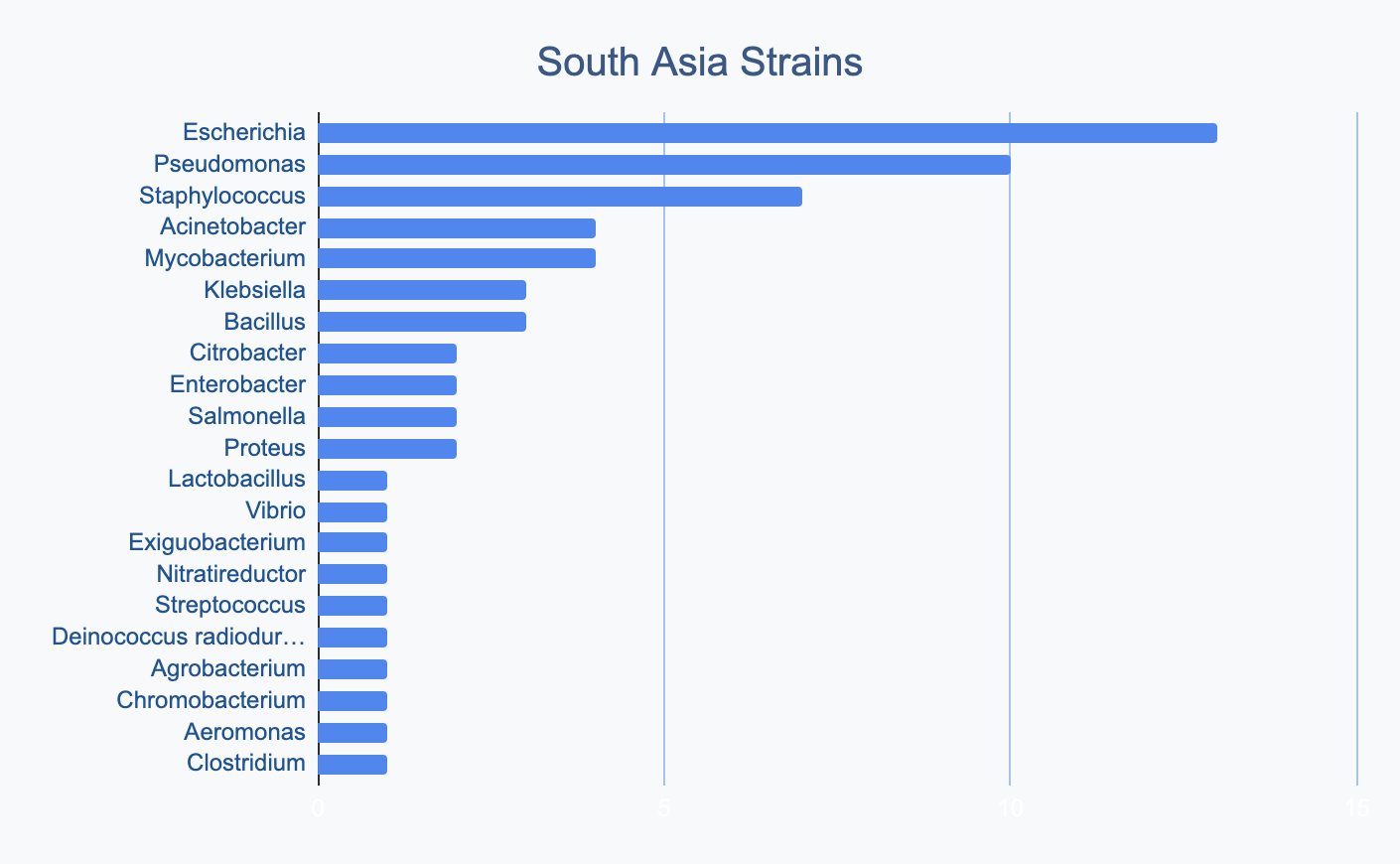
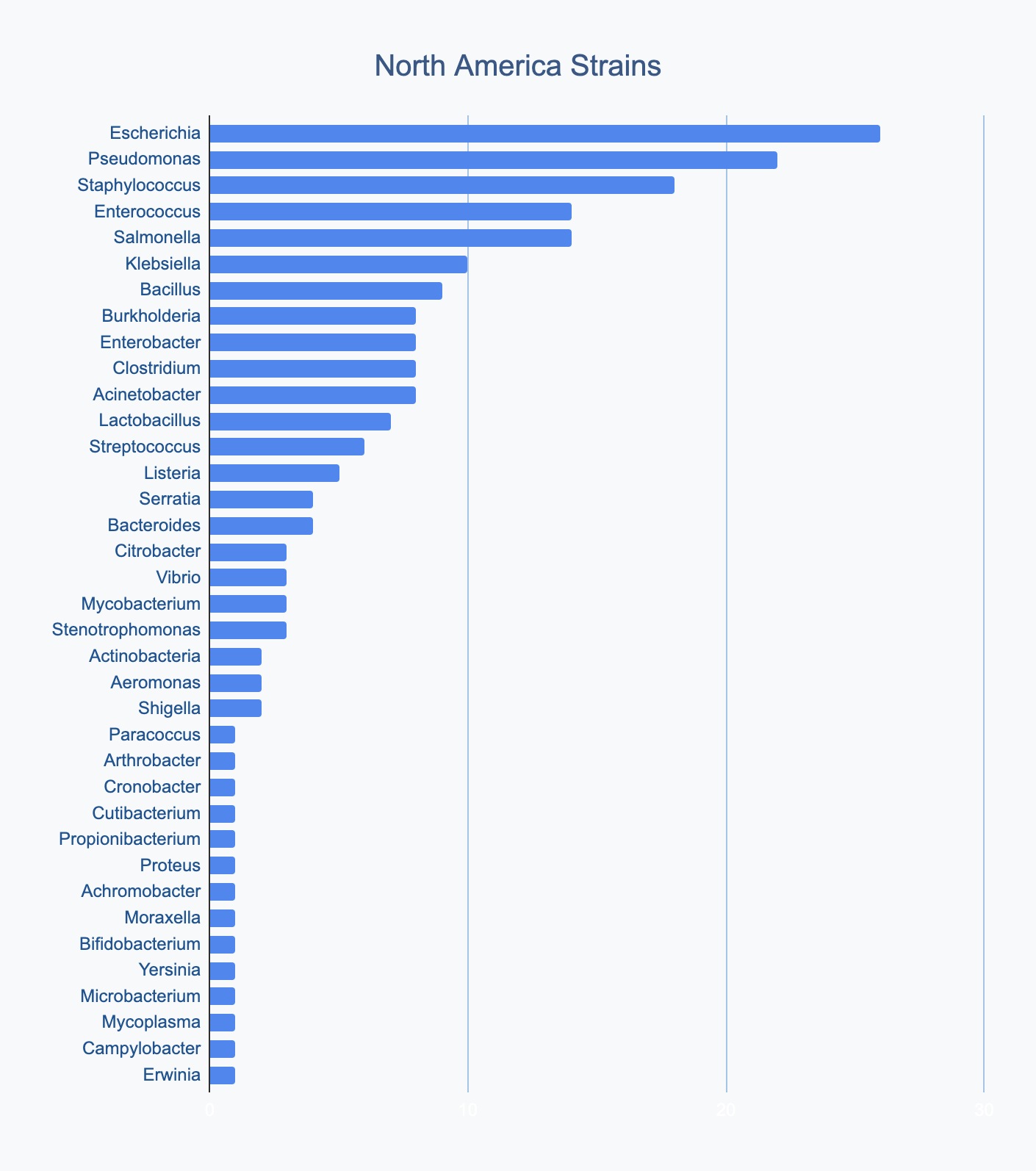
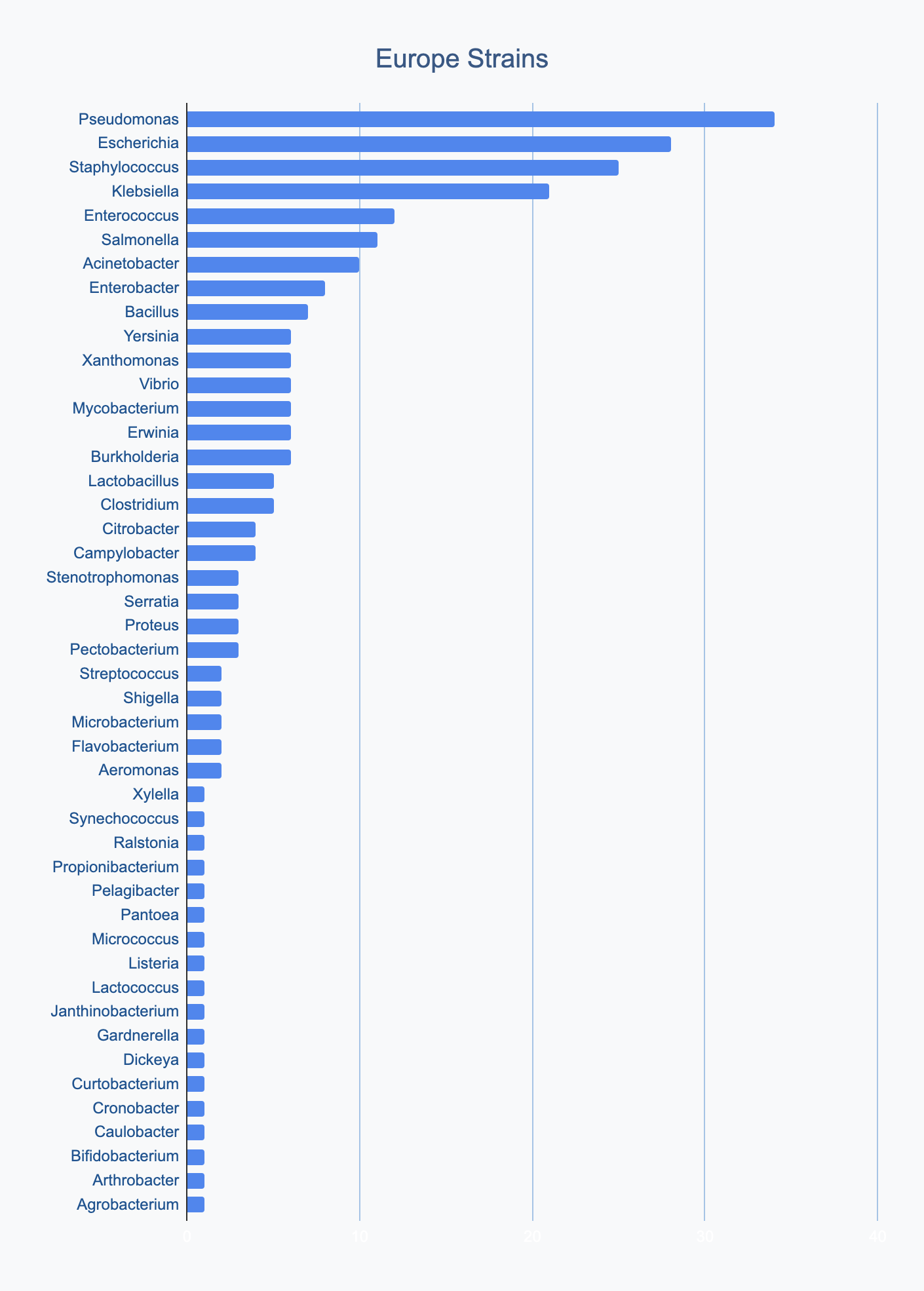
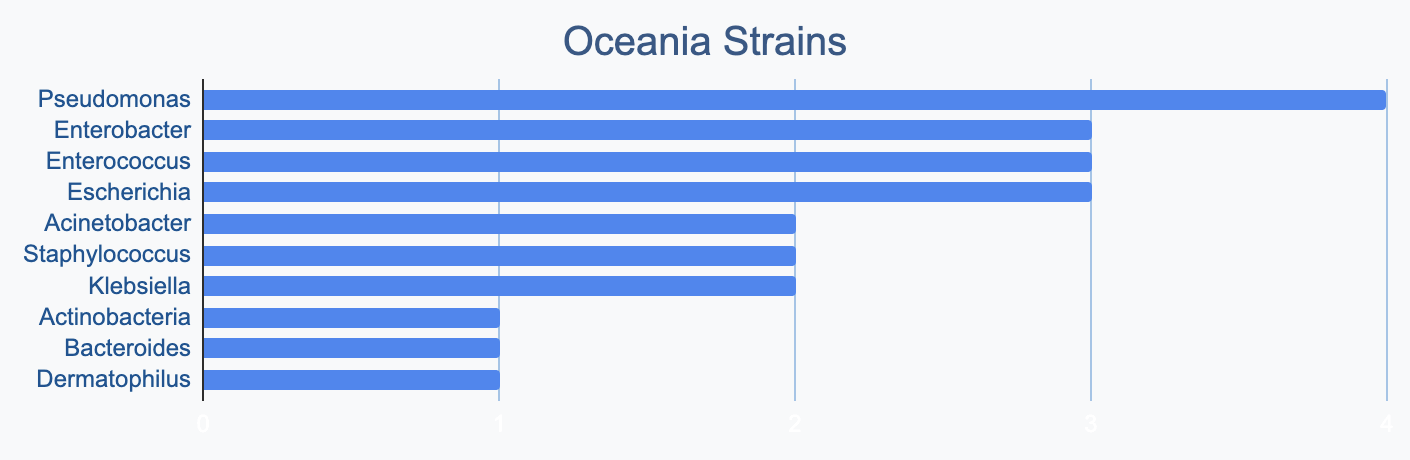
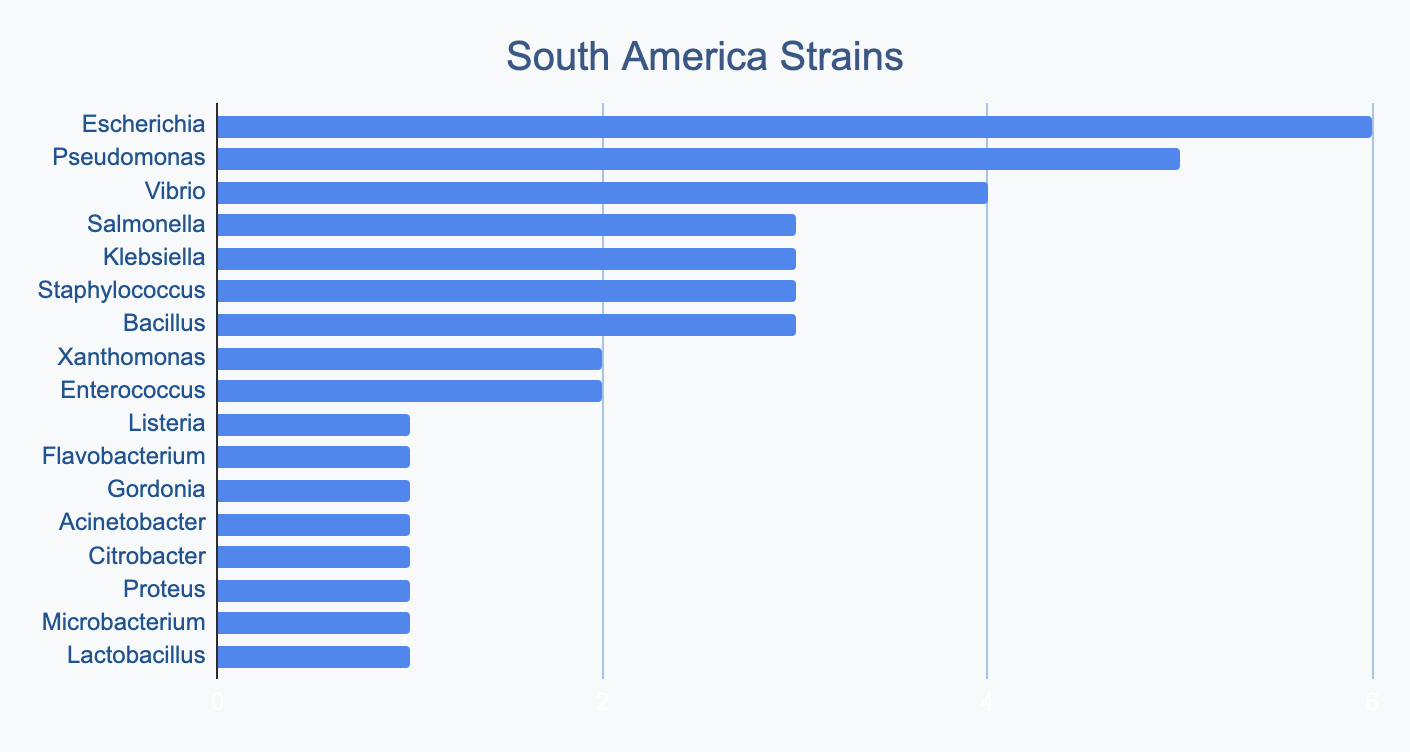
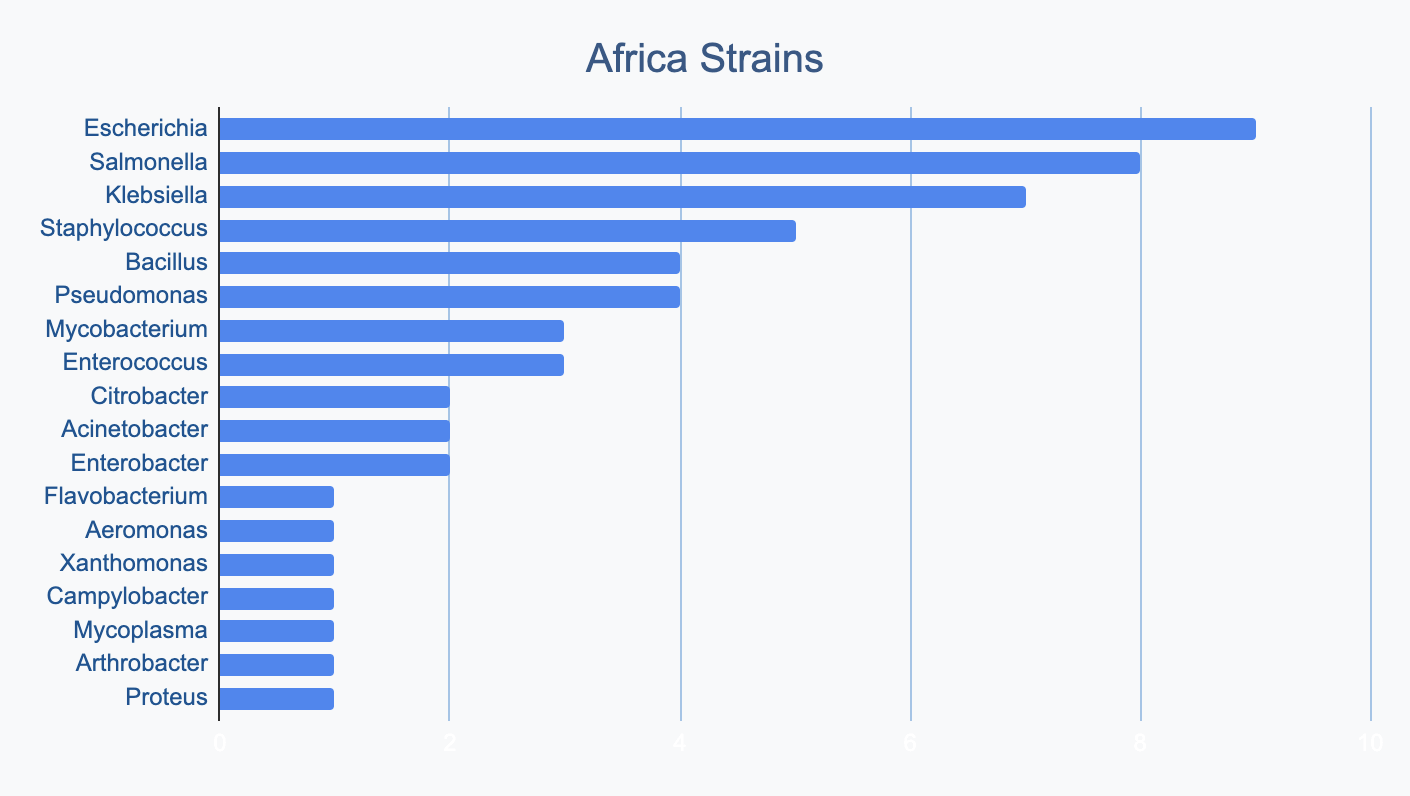
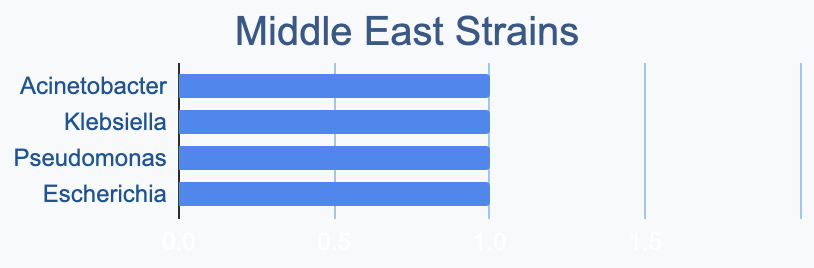
Most labs collect 2-10 different species, with a quarter of labs collecting just one, and a few collecting >10
Looking more closely at the species composition of labs’ strain collections, a quarter of them (~34 labs) only collect one species in their labs. Half of respondents (~70 labs) collect between two and ten species. 15 labs (around 12%) collect more than ten species. 5 labs collect more than 20 species!
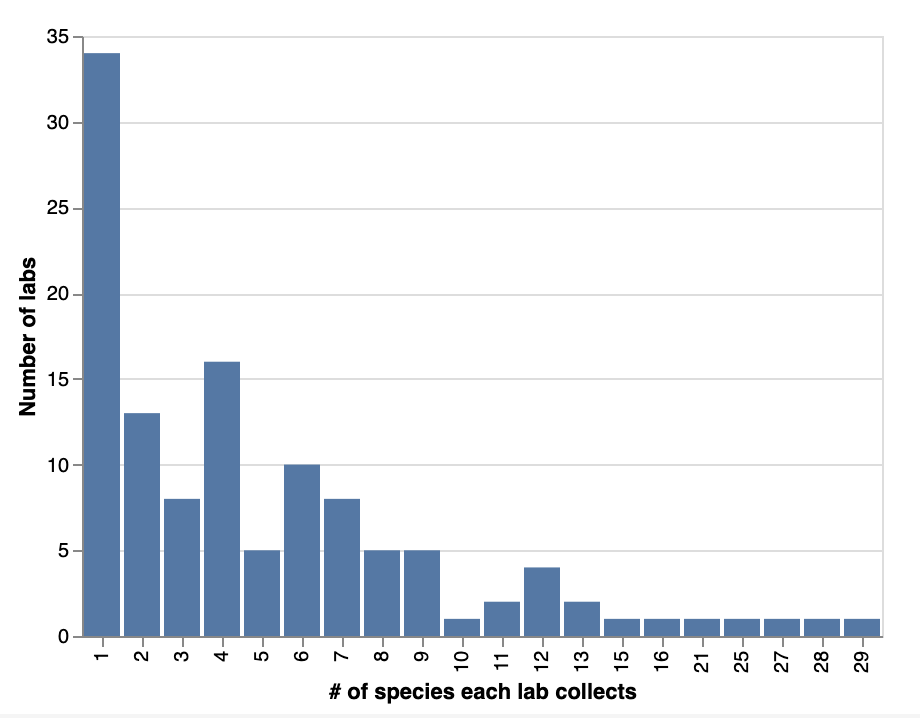
Number of bacterial species that labs collect.
Strain collections come in all sizes, and a third of labs collect at least 200 strains
We asked about the total number of strains, across all species/genera, that each lab collects. All sizes of collections were represented, with similar numbers of respondents reporting collections of under 50 strains as those reporting collections of 50 or more. Surprisingly, 42 labs (1/3) reported they have more than 200 strains in their collections!
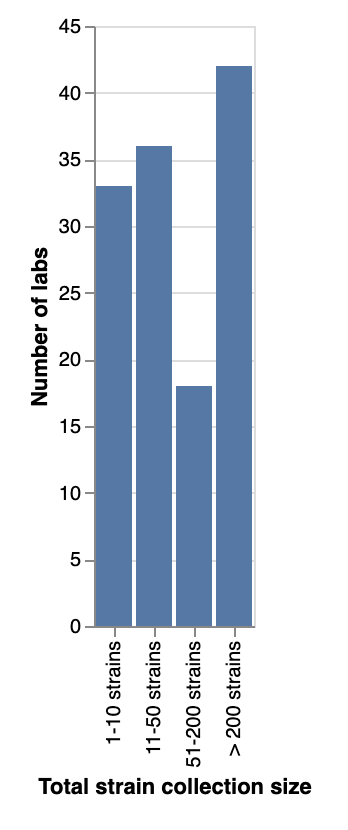
Total number of strains (across all species/genera) that labs collect.
Labs commonly isolate strains themselves or get them from collaborators; few strains came from culture collections
First, most respondents had kept track of how they acquired their strains — few selected ‘unknown’ as a source (we aren’t sure why, but we were assuming we’d see more!). We were surprised that most strains in labs’ collections seem to have been isolated by the lab itself or received from collaborators, rather from culture collections.
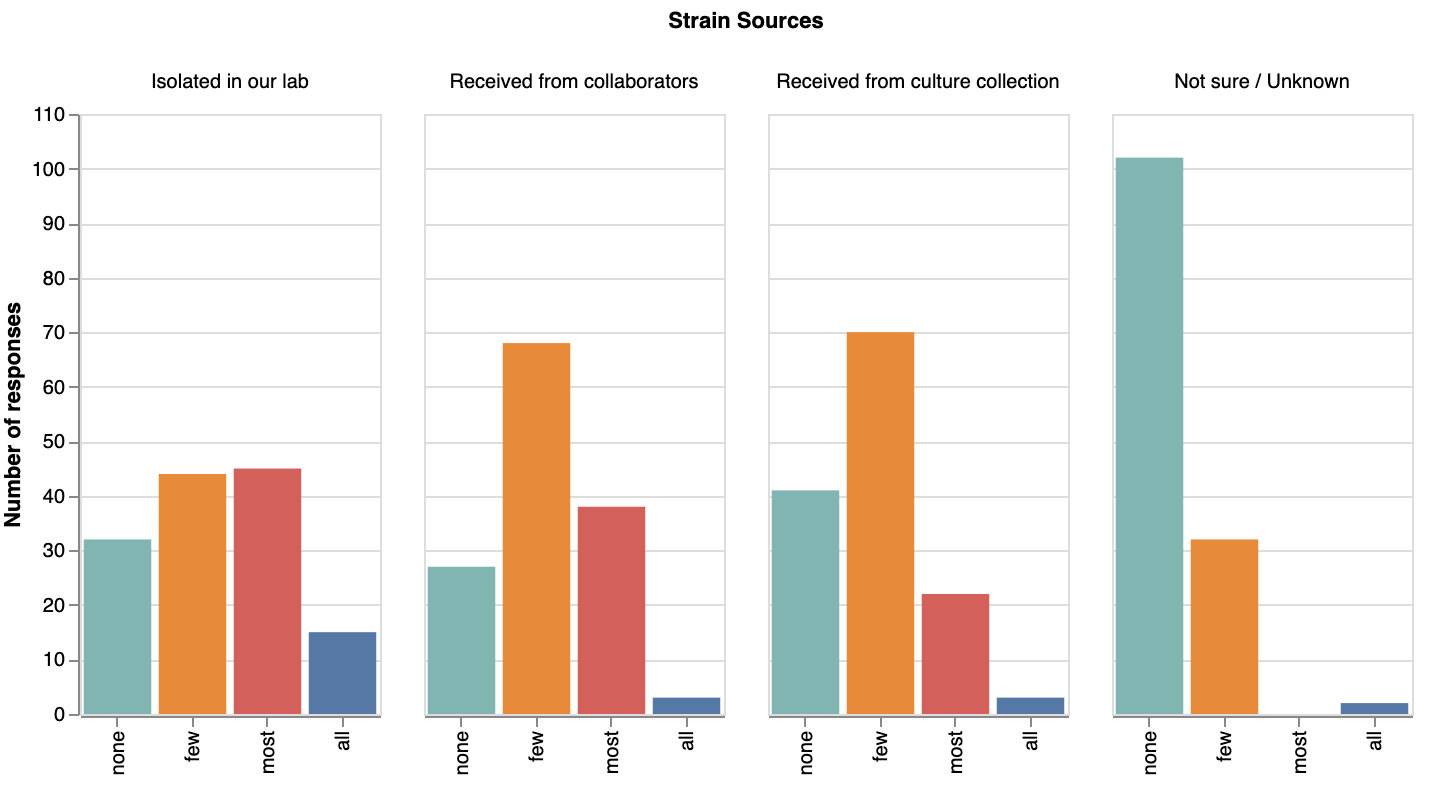
How labs acquired their strains. View larger image
Labs have a good mix of environmental and clinical strains
In terms of original sources of strains, most respondents knew where their strains came from — again, few selected ‘unknown’ as a source. Most strain samples seem to have originated from the environment (though we didn’t ask for more specifics regarding type of environment; now we’re sorry we didn’t!). We were surprised at how many strains came from clinical sources (82 labs reported clinical strains, or 60%), indicating many of our respondents may be working on medically-relevant phage research. Relatively few labs seem to be getting strains from animal sources. Many write-in responses for sources of strains included variations of sewage/waste/feces or water/river/sea/aquatic. A few answers included “plants”. A few unique write-in answers include “Antarctic soil samples”, “insects from windowsill”, “created in our lab”, and “milk of buffalo suffering from mastitis”.
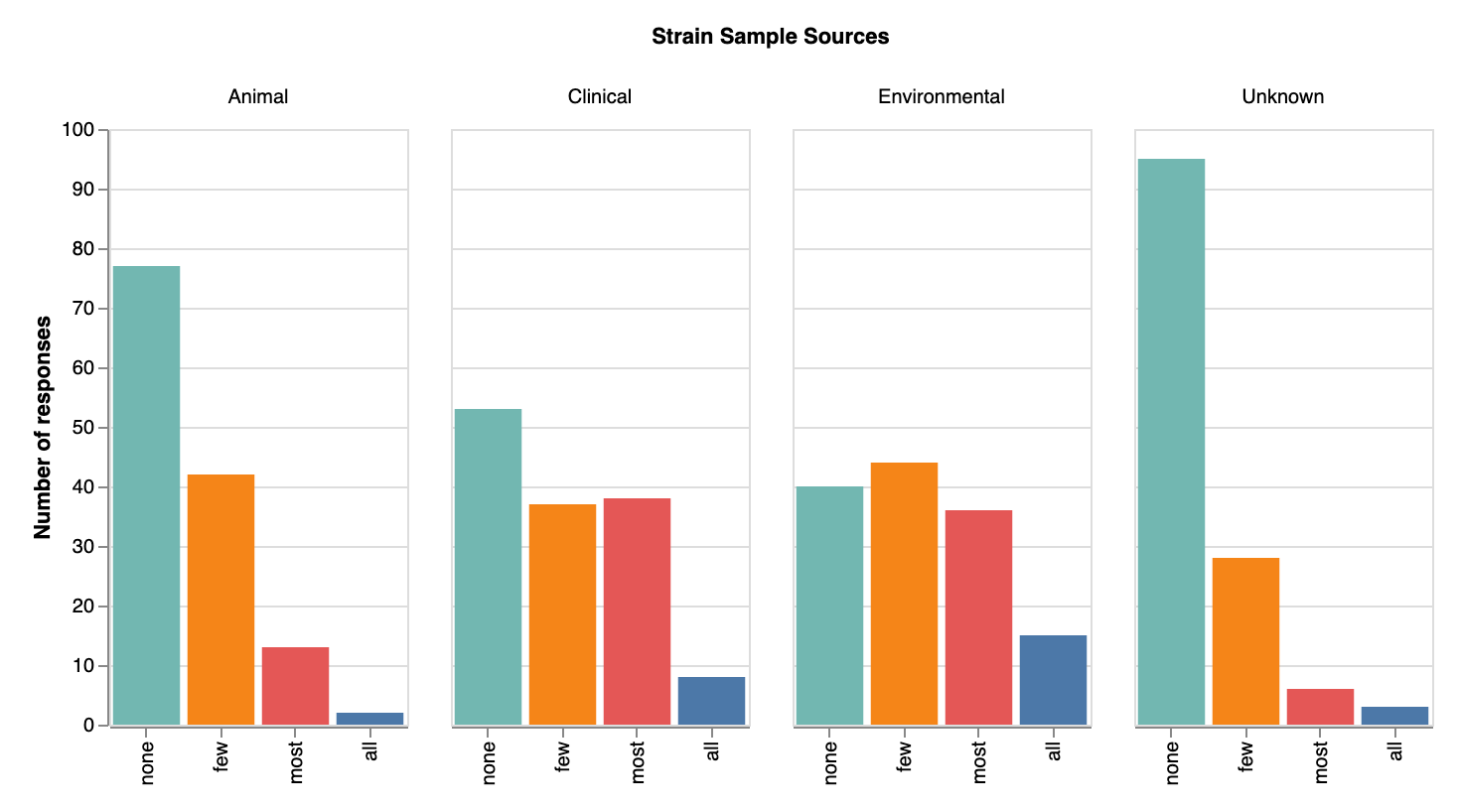
Original sources of strains (across all species/genera) that labs collect. View larger image
A closer look at ESKAPE strain collections
Given the popularity of ESKAPE and clinical strains, we took a closer look at this aspect of the data. We saw that 69 (50.7%) of survey respondents reported having ESKAPE strains*. All ESKAPE pathogens were represented by all collection sizes, suggesting there are large, medium and small collectors of all these pathogens. Acinetobacter, Enterobacter, and Enterococcus were underrepresented in strain collections compared to the more popular Klebsiella, Pseudomonas, and Staphylococcus. It will be interesting to see whether this correlates with the number of Acinetobacter, Enterobacter and Enterococcus phages we see (coming soon in part 3 of our survey results!).
*Note on ESKAPE strains: Tallying up ESKAPE strains accurately was a bit challenging, since the “ESKAPE” terminology combines species like “Klebsiella pneumoniae” and genera “Enterobacter spp”. A limitation of the survey is that we didn’t enforce reporting at the species level, so someone who reported “Klebsiella spp.” may have been using shorthand to refer to Klebsiella pneumoniae, or they may have been reporting a different Klebsiella species that is not considered an ESKAPE strain. Because of this limitation, we decide to look at ESKAPE genera only, meaning that we are including some non-ESKAPE species within these genera as well.
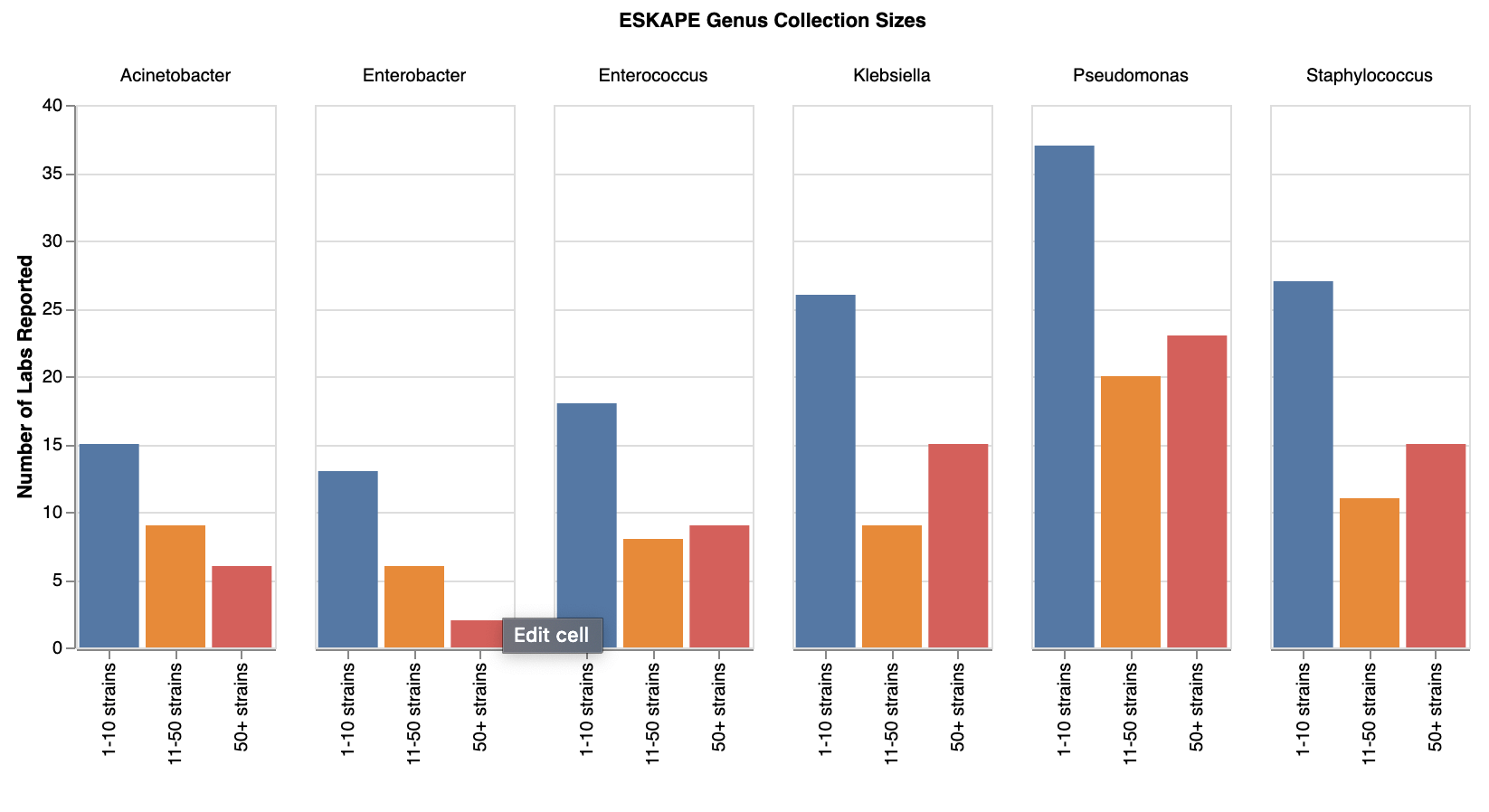
Number of labs with different sizes of ESKAPE strain collections. View larger image
Conclusions
All in all, we saw that labs are collecting a wide array of bacterial species; that each continent has its own fingerprint when it comes to commonly collected strains; that most labs know the sources of their strains, and that they usually isolate strains themselves or get them from collaborators rather than culture collections; that labs have healthy mixes of environmental and clinical strains; and that ESKAPE strains are commonly isolated, with collections of each being gathered on small, medium and large scales.
How we generated the data
Jan used Observable to crunch the numbers, and used Google Sheets and Vega-Lite to generate the graphs.
Your feedback is welcome!
What are you surprised by from this issue? Did we miss anything? We’d love to hear your thoughts! Email [email protected] anytime!
Looking ahead to part 3
In the next State of Phage Snapshot issue, we’ll delve into phage collections. Given the wide range of strains collected, we’re really curious about how big the phage collections are that correspond to the strain collections reported.
Thanks to Sheetal Patpatia for her work writing summaries for the What’s New section this week!






















